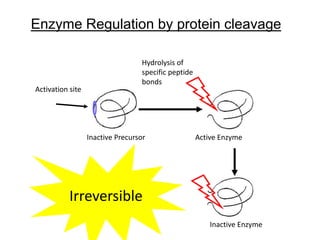
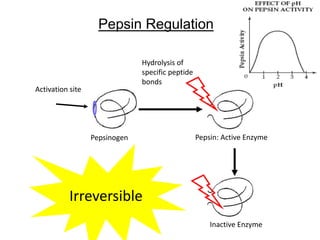
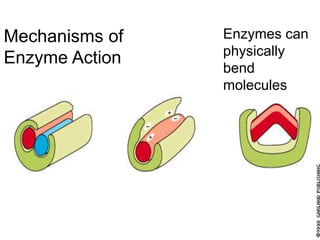
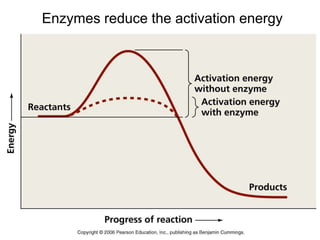
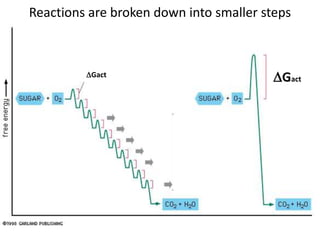
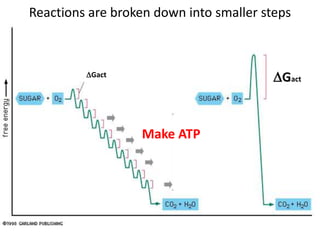
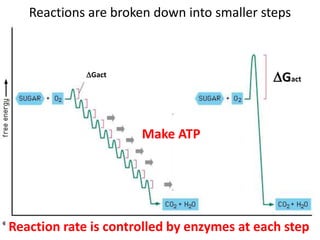
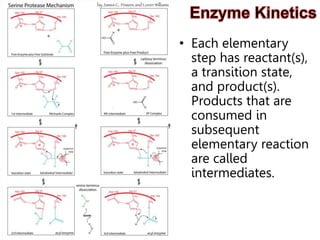
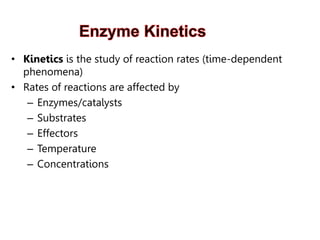
Enzymes catalyze biochemical reactions and lower their activation energy. They do this by holding molecules together, moving electrons between them, and physically manipulating their structures. Enzymes control reaction rates by forming enzyme-substrate complexes at each step. Kinetic studies examine how factors like temperature, concentrations, and effectors influence reaction rates. The Michaelis-Menten model describes how enzyme velocity increases with substrate concentration until plateauing at maximum velocity. Key parameters are KM, the substrate concentration at half-maximal velocity, and kcat, the enzyme's turnover number. Enzyme inhibitors are used as drugs and studied to understand regulation.



![15
Reaction Rates (reaction velocities): To
measure a reaction rate we monitor the
disappearance of reactants or appearance of
products.
e.g., 2NO2 + F2 → 2NO2F
initial velocity =>
[product] = 0,
no back reaction](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-11-320.jpg)

![17
Use experimental data to determine the
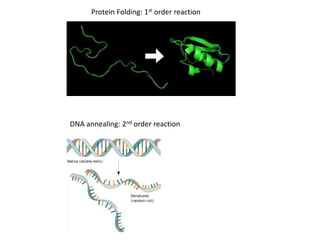
reaction order.
If a plot of [A] vs t is a straight line, then the reaction is zero order.
If a plot of ln[A] vs t is a straight line, then the reaction is 1st order.
If a plot of 1/ [A] vs t is a straight line, then the reaction is 2nd order.
First Order Reaction
-6
-5
-4
-3
-2
-1
0
0.0E+00 2.0E+04 4.0E+04 6.0E+04
Time (in seconds)
ln
[A}
(in
mol
/
L
Second Order Reaction
0
50
100
150
200
250
0 500 1000
Time (in seconds)
1/[A]
(L
/mol)](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-13-320.jpg)

![General Observations
• Enzymes are able to exert their influence at
very low concentrations ~ [enzyme] = nM
• The initial rate (velocity) is linear with
[enzyme].
• The initial velocity increases with [substrate]
at low [substrate].
• The initial velocity approaches a maximum at
high [substrate].](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-18-320.jpg)
![Initial velocity
The initial velocity increases with [S] at low [S].](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-19-320.jpg)
![The initial velocity approaches
a maximum at high [S].
The initial velocity increases with [S] at
low [S].
[velocity =d[P]/dt, P=product]](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-20-320.jpg)
![• Start with a mechanistic model
• Identify constraints and assumptions
• Do the algebra ...
• Solve for velocity (d[P]/dt)](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-21-320.jpg)
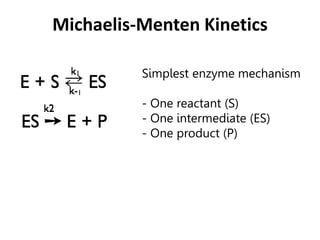
![1. First step: The enzyme (E) and the
substrate (S) reversibly and quickly
form a non-covalent ES complex.
2. Second step: The ES complex
undergoes a chemical
transformation and dissociates to
give product (P) and enzyme (E).
3. v=k2[ES]
4. Many enzymatic reactions follow
Michaelis–Menten kinetics, even
though enzyme mechanisms are
always more complicated than the
Michaelis–Menten model.
5. For real enzymatic reactions use kcat
instead of k2
.](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-23-320.jpg)

![E + S ES E + P
• The enzyme is either free ([E]) or bound ([ES]): [Eo] = [ES] + [E].
• At sufficiently high [S] all of the enzyme is tied up as ES (i.e., [Eo] ≈ [ES],
according to Le Chatelier's Principle)
• At high [S] the enzyme is working at full capacity (v=vmax).
• The full capacity velocity is determined only by kcat and [Eo].
• kcat = turnover #: number of moles of substrate produced per time per
enzyme active site.
kcat
velocity = v =
d[P]
dt
= kcat [ES]
vmax = kcat [E0 ]
kcat =
vmax
[E0 ]
kcat and the reaction velocity
Michaelis-Menten Kinetics](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-25-320.jpg)

![Assumptions
1. k1,k-1>>k2 (i.e., the first step is fast and is
always at equilibrium).
2. d[ES]/dt ≈ 0 (i.e., the system is at steady
state.)
3. There is a single reaction/dissociation step
(i.e., k2=kcat).
4. STot = [S] + [ES] ≈ [S]
5. There is no back reaction of P to ES (i.e. [P] ≈
0). This assumption allows us to ignore k-2. We
measure initial velocities, when [P] ≈ 0.
d[ES]
dt
= rate of formation of ES -
rate of breakdown of ES
» 0 (at steady state)
Michaelis-Menten Kinetics](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-27-320.jpg)
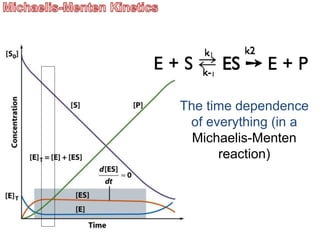
![Now: we derive the Michaelis-Menten Equation
d[ES]/dt = k1[E][S] –k-1[ES] – k2[ES] (eq 12-14 VVP)
= 0 (steady state assumption, see previous graph)
solve for [ES] (do the algebra)
[ES] = [E][S] k1/(k-1 + k2)
Define KM (Michealis Constant)
KM = (k-1 + k2)/k1 => [ES] = [E][S]/KM
rearrange to give KM = [E][S]/[ES]](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-29-320.jpg)
![substitute [E]=[E]0 -[ES]
([E]0 -[ES])[S]
[ES]
= KM eqs 12-20 VVP
multiply both sides by [ES]
KM [ES]= ([E]0 -[ES])[S]
solve for [ES]
[ES]=
[E]0[S]
Km +[S]
eq 12-22 VVP
multiply both sides by k2 (this gives get the velocity of the reaction)
dP
dt
= v = k2[ES]=
k2[E]0[S]
KM +[S]
eq 12-23 VVP
and remember that k2[E]0 = vmax
v =
vmax[S]
KM +[S]
Michaelis Menten Equation eq 12-25 VVP
KM = [E][S]/[ES]](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-30-320.jpg)
![v =
vmax[S]
KM +[S]
Michaelis Menten Equation eq 12-25 VVP
When [S] = KM then,
v =
vmax[S]
[S]+[S]
=
vmax
2
This is saying that when KM =[S], the reaction runs at half maximum velocity.](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-31-320.jpg)
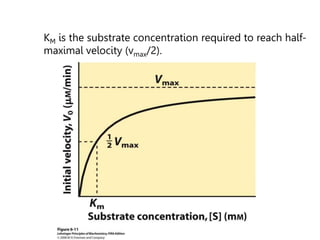
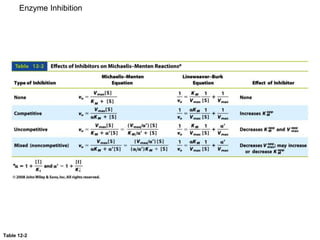
![Significance of KM
• KM = [E][S]/[ES] and KM = (k-1 + k2)/k1.
• KM is the apparent dissociation constant of the ES complex. A
dissociation constant (KD) is the reciprocal of the equilibrium constant
(KD=KA
-1). KM is a measure of a substrate’s affinity for the enzyme (but
it is the reciprocal of the affinity).
• If k1,k-1>>k2, the KM=KD.
• KM is the substrate concentration required to reach half-maximal
velocity (vmax/2). A small KM means the sustrate binds tightly to the
enzyme and saturates (max’s out) the enzyme.
• The microscopic meaning of Km depends on the details of the
mechanism.](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-33-320.jpg)
![Significance of kcat/KM
• kcat/KM is the catalytic efficiency. It is used to rank enzymes. A big
kcat/KM means that an enzyme binds tightly to a substrate (small KM),
with a fast reaction of the ES complex.
•
• kcat/KM is an apparent second order rate constant
v=kcat/KM[E]0[S]
• kcat/KM can be used to estimate the reaction velocity from the total
enzyme concentration ([E]0). kcat/KM =109 => diffusion control.
• kcat/KM is the specificity constant. It is used to distinguish and describe
various substrates.](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-35-320.jpg)
![Data analysis
• It would be useful to have a linear plot of
the MM equation
• Lineweaver and Burk (1934) proposed the
following: take the reciprocal of both
sides and rearrange.
• Collect data at a fixed [E]0.](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-36-320.jpg)
![v =
vmax[S]
KM +[S]
Michaelis Menten Equation eq 12-25
take the reciprocal
1
v
=
KM +[S]
vmax[S]
=
KM
vmax[S]
+
1
vmax
Graph
1
v
versus
1
[S]
the y (1/v) intercept (1/[S] = 0) is 1/vmax
the x (1/[S]) intercept (1/v = 0) is -1/KM
the slope is KM/vmax](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-37-320.jpg)
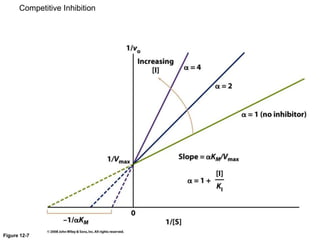
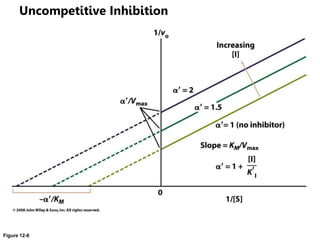
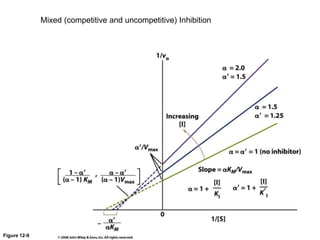
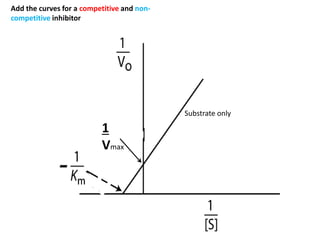
![Lineweaver-Burk-Plot
the y (1/v) intercept (1/[S] = 0) is 1/vmax
the x (1/[S]) intercept (1/v = 0) is -1/KM
the slope is KM/vmax](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-38-320.jpg)
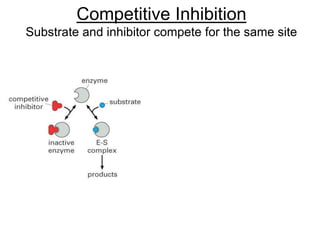
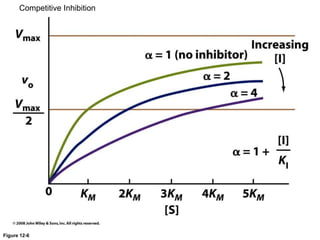
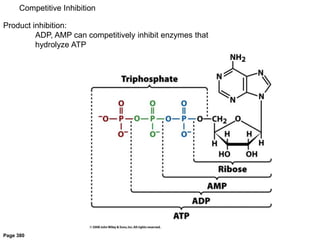
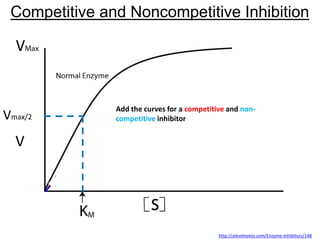
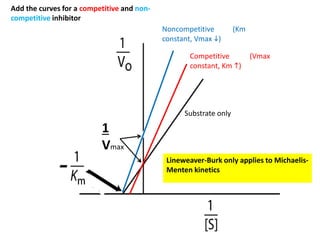
![Competitive Inhibition
Substrate and inhibitor compete for the same site
If [S]>>[I]
Vmax is the same, Km h
Vmax
1/Km](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-44-320.jpg)
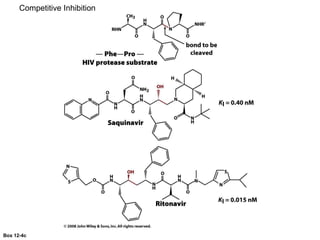
![Competitive Inhibition
Substrate and inhibitor compete for the same site
If [S]>>[I]
Vmax is the same, Km h
Vmax
What happens to the slope? Vmax? Km?](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-45-320.jpg)
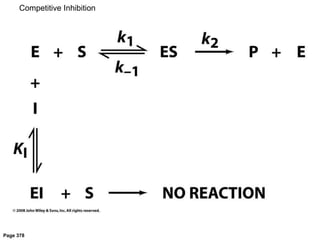
![[E][I]
[EI]
= KI inhibitor dissociation constant
[E]0 =[E] -[ES]-[EI] total enzyme concentration
a =1+
[I]
KI
1
v
=
aKM +[S]
vmax[S]
=
aKM
vmax[S]
+
1
vmax
Competitive Inhibition](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-47-320.jpg)
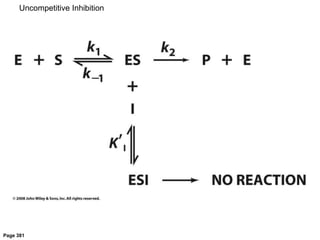
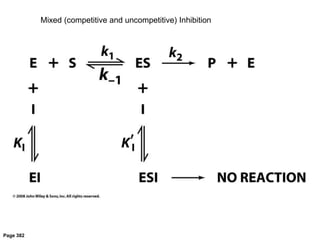
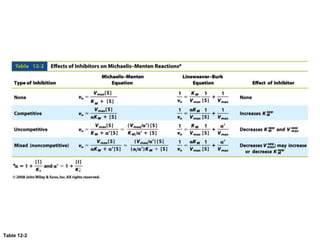
![[ES][I]
[ESI]
= KI inhibitor dissociation constant
[E]0 =[E] -[ES]-[ESI] total enzyme concentration
1
v
=
KM
vmax[S]
+
a
vmax
Uncompetitive Inhibition](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-53-320.jpg)
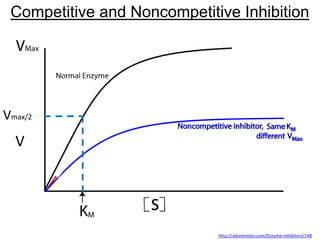
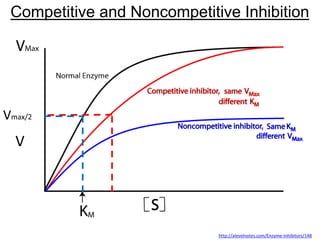
![Noncompetitive
Inhibition
What happens to Vmax & Km?
a. Vmax same, Kmh
b. Vmax same, Kmi
c. Vmaxh, Km same
d. Vmaxi , Km same
X
http://en.wikipedia.org/wiki/Non-competitive_inhibition#mediaviewer/File:Non-competitive_inhibition.svg
No inhibitor
EI cannot be used;
Vmax = k2[E]with no inhibitor](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-58-320.jpg)
![Task:For an enzyme that follows Michaelis-Menten kinetics, by
what factor does the substrate concentration need to increase
to change the rate of the reaction from 20% to 80% Vmax?
]
[
]
[
max
S
K
S
V
V
m
A. two-fold
B. 4-fold
C. 8-fold
D. 16-fold
E. 24-fold](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-64-320.jpg)
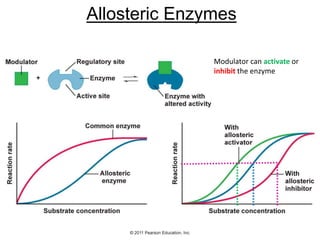
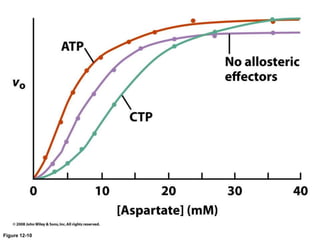
![Shift at physiological concentrations of substrate
Allosteric
“T” form
Slight h[S] (in pink region) causes transition from “T” to “R”
Michaelis-Menten
“R” form](https://image.slidesharecdn.com/enzymes-bme-230925124208-761b21d0/85/ENZYMES-bme-pptx-69-320.jpg)