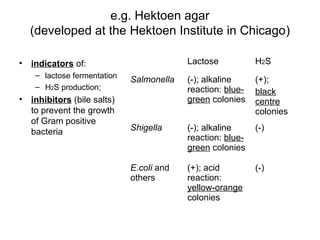
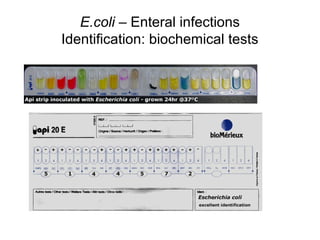
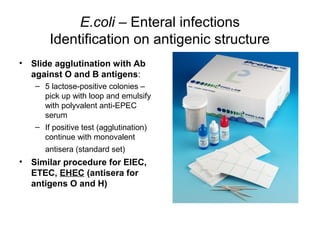
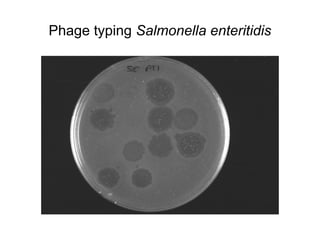
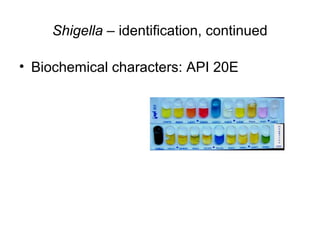
This document provides information on laboratory diagnosis of infections caused by the family Enterobacteriaceae. It discusses the collection and testing of various specimen types, including stool, urine, and blood. A variety of culture media and biochemical tests are described to isolate and identify bacterial pathogens like E. coli, Salmonella, and Shigella from clinical samples. Identification methods include antigenic structure identification using agglutination tests and phage typing for certain Salmonella serotypes. The document provides details on laboratory diagnosis of common infections like urinary tract infections and enteric diseases including typhoid fever.