DNA replication, repair and recombination Notes
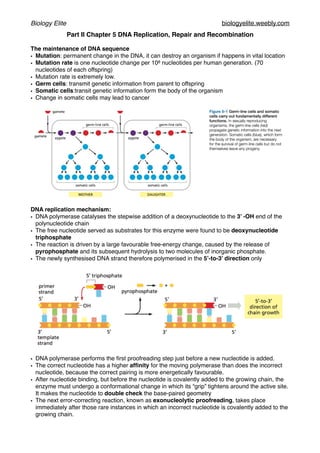
- 1. Biology Elite biologyelite.weebly.com Part II Chapter 5 DNA Replication, Repair and Recombination The maintenance of DNA sequence • Mutation: permanent change in the DNA, it can destroy an organism if happens in vital location • Mutation rate is one nucleotide change per 108 nucleotides per human generation. (70 nucleotides of each offspring) • Mutation rate is extremely low. • Germ cells: transmit genetic information from parent to offspring • Somatic cells:transit genetic information form the body of the organism • Change in somatic cells may lead to cancer DNA replication mechanism: • DNA polymerase catalyses the stepwise addition of a deoxynucleotide to the 3’ -OH end of the polynucleotide chain • The free nucleotide served as substrates for this enzyme were found to be deoxynucleotide triphosphate • The reaction is driven by a large favourable free-energy change, caused by the release of pyrophosphate and its subsequent hydrolysis to two molecules of inorganic phosphate. • The newly synthesised DNA strand therefore polymerised in the 5’-to-3’ direction only • DNA polymerase performs the first proofreading step just before a new nucleotide is added. • The correct nucleotide has a higher affinity for the moving polymerase than does the incorrect nucleotide, because the correct pairing is more energetically favourable. • After nucleotide binding, but before the nucleotide is covalently added to the growing chain, the enzyme must undergo a conformational change in which its “grip” tightens around the active site. It makes the nucleotide to double check the base-paired geometry • The next error-correcting reaction, known as exonucleolytic proofreading, takes place immediately after those rare instances in which an incorrect nucleotide is covalently added to the growing chain.
- 2. Biology Elite biologyelite.weebly.com • DNA molecules with a mismatched (improperly base-paired) nucleotide at the 3ʹ-OH end of the primer strand are not effective as templates because the polymerase has difficulty extending such a strand. • DNA polymerase in this case will turn into editing mode, turning into 3’-to-5’ proofreading exonuclease clipping off any unpaired or misfired nucleotides. • DNA polymerase functions as a “self-correcting” enzyme that removes its own polymerisation errors as it moves along the DNA • If there were a DNA polymerase that added deoxyribonucleotide triphosphates in the 3ʹ-to-5ʹ direction, would have to provide the activating triphosphate needed for the covalent linkage. In this case, the mistakes in polymerisation could not be simply hydrolysed away, because the bare 5ʹ end of the chain thus created would immediately terminate DNA synthesis. • Only DNA replication in the 5’-to-3’ direction allows efficient error correction • DNA primases adds a RNA primer on the lagging strand for about 10 nucleotides long at the intervals of 100-200 nucleotides • Any enzyme that starts a new chain cannot be self-correct. So reason of using a RNA primer instead of a DNA primer is to keep the accuracy of replication. DNA primer may make mistakes without self-correcting mechanism. RNA primer will be efficiently removed ad replaced.
- 3. Biology Elite biologyelite.weebly.com • DNA helicases were first isolated as proteins that hydrolysed ATP when they are bound to single strand of DNA. The hydrolysis of ATP can change the shape of a protein molecule in a cyclical manner that allows the protein to perform mechanical work. • Single-strand DNA-binding (SSB) proteins, also called helix-destabilizing proteins, bind tightly and cooperatively to exposed single-strand DNA without covering the bases, which therefore remain available as templates. These proteins are unable to open a long DNA helix directly, but they aid helicases by stabilising the unwound, single-strand conformation. • On their own, most DNA polymerase will synthesis only a short distance before falling off. This property allows DNA polymerase to be recycled so quickly on the lagging strand • A sliding clamp helps the DNA polymerase sticks firmly on the DNA while moving. The clamp is a type of accessory protein (PCNA for eukayotes) • The clamp has a large ring shape around the DNA double helix. The assembly of the clamp requires ATP hydrolysis by a special protein, called clamp loader, which hydrolyses ATP as it loads the clamp on to a primer-templates junction. • In eukaryotes, Polδ completes each Okazaki fragment on the lagging strand and Polε extends the leading strand. • Summary: At the front of the replication fork, DNA helicase opens the DNA helix. Two DNA polymerase molecules work at the fork, one on the leading strand and one on the lagging strand. Whereas the DNA polymerase molecule on the leading strand can operate in a continuous fashion, the DNA polymerase molecule on the lagging strand must restart at short
- 4. Biology Elite biologyelite.weebly.com intervals, using a short RNA primer made by a DNA primase molecule. The close association of all these protein components increases the efficiency of replication and is made possible by a folding back of the lagging strand. This arrangement also facilitates the loading of the polymerase clamp each time that an Okazaki fragment is synthesised: the clamp loader and the lagging-strand DNA polymerase molecule are kept in place as a part of the protein machine even when they detach from their DNA template. • Strand-directed mismatch repair system detects the potential for distortion in the DNA helix from the misfit between non complementary base pairs. • In E.coli, methylation of all A residues in the sequence GATC is used to distinguish between the old strand and the newly made strand. • In eukaryotes, newly synthesized lagging-strand DNA transiently contains nicks (before they are sealed by DNA ligase) and such nicks (also called single-strand breaks) provide the signal that directs the mismatch proofreading system to the appropriate strand. • Having a defective copy of mismatch repair genes in human can be very dangerous, it may lead to hereditary nonpolyposis colon cancer (HNPCC) due to rapid accumulation of mutations. • As replication fork moves along the DNA double helix, it creates a winding problem. For every 10 pairs of nucleotides being replication, one turn of the DNA helix must be completed. However, turing the helix is energetically unfavourable, which will create overwound of DNA in front of replication fork. • DNA topoisomerase is used to solve this problem. It can be viewed as a reversible nuclease that adds itself covalently to DNA backbone, breaking phosphdiester bond. The reaction is reversible.
- 5. Biology Elite biologyelite.weebly.com • Topoisomerase I produces a transient single-strand break, which allows two sections of DNA rotates freely to each other. • Topoisomerase II produces a transient double-strand break, it breaks one double helix reversibly to create a gate, inducing a nearby DNA helix to pass through the gate and then closes it.
- 6. Biology Elite biologyelite.weebly.com The initiation and completion of DNA replication in chromosomes • DNA replication begins at replication origin, where is rich of A-T base pairs due to weaker forces between two strands (2 hydrogen bonds) • Bacteria only has a single origin of DNA replication • Replication origins attract initiator proteins that binds to double-stranded DNA and pull it apart • In human, replication of average-sized chromosome will take 35 days if there is only a single replication origin. In eukaryotes, there are multiple replication origin. • DNA sequences that can serve as an origin of replication are found to contain: binding site for a large, multisubunit initiator protein called ORC (origin recognition complex), a stretch of DNA that is rich in As and Ts, at least one binding site for protein that facilitate ORC. • In brief, during G1 phase, the replicative helicases are loaded onto DNA next to ORC to create a prereplicative complex. Then, upon passage from G1 phase to S phase, specialized protein kinases come into play to activate the helicases. The resulting opening of the double helix allows the loading of the remaining replication proteins, including the DNA polymerases. The protein kinases that trigger DNA replication simultaneously prevent assembly of new prereplicative complexes until the next M phase resets the entire cycle • This strategy provides a single window of opportunity for prereplicative complexes to form; thus ensure all DNA is copied once only. • Histone proteins are required to package DNA and they are usually made only in the S phage during DNA replication.
- 7. Biology Elite biologyelite.weebly.com • When a nucleosome is traversed by a replication fork, the histone octamer appears to be broken into an H3-H4 tetramer and two H2A-H2B dimers • The H3-H4 tetramer remains loosely associated with DNA and is distributed at random to one or the other daughter duplex, but the H2A-H2B dimers are released completely from DNA. • Freshly made H3-H4 tetramers are added to the newly synthesized DNA to fill into the “spaces,” and H2A-H2B dimers—half of which are old and half new—are then added at random to com- plete the nucleosomes • As DNA polymerase δ discontinuously synthesizes the lagging strand, the length of each Okazaki fragment is determined by the point at which DNA polymerase δ is blocked by a newly formed nucleosome. • This explains why the length of Okazaki fragments in eukaryotes (~200 nucleotides) is approximately the same as the nucleosome repeat length. • The orderly and rapid addition of new H3-H4 tetramers and H2A-H2B dimers behind a replication fork requires histone chaperones (also called chromatin assembly factors). • The histone chaperones, along with their cargoes (histone proteins), are directed to newly replicated DNA through a specific interaction with the eukaryotic sliding clamp called PCNA • These clamps are left behind moving replication forks and remain on the DNA long enough for the histone chaperones to complete their tasks. • When the replication fork reaches an end of a linear chromosome, the final RNA primer synthesised on the lagging strand cannot be replaced by DNA because there is no 3’-OH end available for repair polymerase, which means DNA will lost a part of its end during every DNA replication • In bacteria, circular DNA solves this problem. In eukaryotes, specialised nucleotide sequence at the end of the chromosomes called telomeres can solved this problem. Telomeres contain many tandem repeat, in human is GGGTTA • Telomere DNA sequences are recognized by sequence-specific DNA-binding proteins that attract an enzyme, called telomerase
- 8. Biology Elite biologyelite.weebly.com • Telomerase recognises the tip of an existing telomere DNA repeat sequence and elongates it in the 5ʹ-to-3ʹ direction, using an RNA template that is a component of the enzyme itself to synthesise new copies of the repeat • Telomeres must clearly be distinguished from these accidental breaks; otherwise the cell will attempt to “repair” telomeres, causing chromosome fusions and other genetic abnormalities. • A specialized nuclease chews back the 5ʹ end of a telomere leaving a protruding single-strand end. • This protruding end—in combination with the GGGTTA repeats in telomeres—attracts a group of proteins that form a protective chromosome cap known as shelterin. In particular, shelterin “hides” telomeres from the cell’s damage detectors that continually monitor DNA. DNA Repair: • Defeats in human repair genes can lead to some diseases due to high mutation rate such as hereditary cool cancer, breast cancer and xeroderma pigmentosum (XP) • DNA double helix can be damaged in many ways, including deprivation (loss of guanine), deamination (cytosine to uracil), reactive metabolites, chemicals in the environment and radiation.
- 9. Biology Elite biologyelite.weebly.com Base excision repair: specific base change • DNA glycosylase: recognise a specific type of altered base in DNA and catalyse its hydrolytic removal, including: those that remove deaminated Cs, deaminated As, different types of alkylated or oxidized bases, bases with opened rings, and bases in which a carbon–carbon double bond has been accidentally converted to a car- bon–carbon single bond • AP endonuclease: cut the sugar backbone and add in nucleotides. Depurination can be therefore directly repaired by AP endonuclease. Nucleotide excision repair: distortion in double helix • Excision nuclease finds out distortion in double helix, including covalent reaction between DNA bases and large hydrocarbons and base dimer (T-T,C-T,C-C) • DNA helicase cuts the section of distorted DNA out • DNA polymerase rebuilds the double helix and DNA ligase ligates the helix • Transcription-coupled excision repair: Nucleotide excision repair protein couples with RNA polymerase which ensures the vital sections of genes are correct in gene expression • Translesion DNA polymerase is used in emergencies for replication highly-damaged DNA sequences. However, they are not accurate as DNA polymerase due to lack of exonucleolytic proofreading activity. They are only released in emergencies and make “good guesses”.
- 10. Biology Elite biologyelite.weebly.com Nonhomologous end joining: • broken ends are simply brought together by DNA ligation • quick and dirty • deletion of DNA sequences occur at the site of ligation and will lead to loss of nucleotides • small amount of nucleotides loss is acceptable in mammalian somatic cells due to a large genome • mistakes can happen: broken chromosomes mistakenly covalently attach to another Homologous recombination: • accurately correct double stranded break • homologous recombination often occurs just after DNA replication, when the two daughter DNA molecules lie close together and one can serve as a template for repair of the other. • 5’ end of the damaged DNA is digested by specialised nuclease to produce overhanging single-strand 3’ end • Strand exchange: one of the single-strand 3ʹ ends from the damaged DNA molecule worms its way into the template duplex and searches it for homologous sequences through base-pairing.
- 11. Biology Elite biologyelite.weebly.com • An accurate DNA polymerase extends the invading DNA strand using the information form the undamaged strand. • invading strand relates and reform the broken double helix. • DNA synthesis continues using the strands from damaged DNA as templates • DNA ligation to form complete double helix. Strand Exchange: • special protein does this job, in E Coli. is RecA and in all eukaryotes is Rad51 • RecA first binds cooperatively to the invading single strand, forming a protein–DNA filament that forces the DNA into an unusual configuration: groups of three consecutive nucleotides are held as though they were in a conventional DNA double helix but, between adjacent triplets, the DNA backbone is untwisted and stretched out • This unusual protein–DNA lament then binds to duplex DNA in a way that stretches the duplex, destabilizing it and making it easy to pull the strands apart. • The invading single strand then can sample the sequence of the duplex by conventional base-pairing. This sampling occurs in triplet nucleotide blocks: if a triplet match is found, the adjacent triplet is sampled, and so on.
- 12. Biology Elite biologyelite.weebly.com • Homologous recombination can also repair replication fork. • Replication fork may fall off due to nick or a gap in the parental DNA helix just ahead the replication fork. Regulate the use of homologous recombination: • sometimes a broken human chromosome is repaired using the homolog from the other parent instead of the sister chromatin • maternal and paternal chromosomes differ in DNA sequence at many positions along their lengths. Homologous recombination can convert the sequence of the repaired DNA from the maternal to the paternal sequence or vice versa. This type of recombination is known as loss of heterozygosity. Homologous Recombination is crucial for meiosis: • programmed double stranded break is preformed by a specialised protein (Spo11 in budding yeast). Like a topoisomerase, Spo11 remains on the broken DNA sequence • Specialised nuclease chews back at the 5’ end of the double helix, degraded the Spo11 and leaving a overhanging 3’ end • Holiday junction (cross-strand exchange) is formed, two double-strand DNA helixes are connected with specific protein, thereby stabilises the open symmetric isomers.
- 13. Biology Elite biologyelite.weebly.com • Specialised proteins that bind to the holiday auctions can catalyse a reaction known as branch migration, whereby DNA is spooled through the holiday auction by continually breaking and reforming. • Holiday auction therefore can move and expand the region of heteroduplex DNA from initial site using the energy from ATP • The outcome of the holiday junction can be non-crossover or crossover. 90% of homologous recombination is non-crossover. But the crossover has significant meanings. • We don’t know what decide crossover to happen. We know that crossover in one position will inhibit crossover in the neighbouring regions. Crossover control ensures the roughly even distribution of crossover points along the chromosomes. • Roughly two crossovers occur per chromosome per mitosis • In both crossover and non-crossover, recombination will leave a heteroduplex region where a strand of paternal DNA is paired with a strand of maternal DNA. The regions can last for thousands of nucleotides due to branch migration.
- 14. Biology Elite biologyelite.weebly.com Gene conversion: If the two strands that make up a heteroduplex region do not have identical nucleotide sequences, mismatched base pairs are formed, and these are often repaired by the cell’s mismatch repair system. However, the mismatch repair system cannot distinguish between the paternal and maternal strands and will randomly choose the strand to be used as a template. As a consequence, one allele will be lost and the other duplicated, resulting in net “conversion” of one allele to the other. Transposition recombination: • Mobile genetic element: a wide variety of specialised segments of DNA that can be moved from one position in a genome into another • Mobile elements that move by the way of transposition are called transposons, or transposable elements • In transposition, a specific enzyme, usually encoded by the transposon itself and typically called a transposase, acts on specific DNA sequences at each end of the transposon, causing it to insert into a new target DNA site. • Most transposons move very rarely, in bacteria, transposons move once per 105 cell division • More frequent movement will probably destroy the cell genome. • Transposons can be classified into DNA-only transposons, retroviral-like retrotransposons, nonretroviral retrotransposons. • DNA-only transposon: they exist only as DNA during their movement, predominate in bacteria and they are largely responsible for the spreading of antibiotic resistance. • DNA-only transposon can be relocated from the donor site to the target site by cut-and- paste transposition. This reaction produces a short duplicated of the target DNA sequence at the insertion site, which makes transposon inserted and ligated perfectly to the insertion site. At both ends of transposon, short inverted repeat sequence are found to indicated its identity. • Double-stranded break cause by the loss of transposons can be repaired either by homologous recombination or non-homologous end joining which will leaves a mutation at the original transposon site.
- 15. Biology Elite biologyelite.weebly.com • Certain viruses are considered mobile genetic elements because they use transposition mechanism to integrate their genomes into that of their host cell. • Retrovirus: exists as a single-stranded RNA genome packed into a protein shell along with a virus-encoded reverse transcriptase enzyme • The infection procedures of retrovirus involves turning single-stranded RNA into double stranded DNA by reverse transcriptase, then virus-encoded transposase called integrase inserts the viral DNA into the chromosome by a cut-and-paste transposition. • Retroviral-like retrotransposons is relocated like retrovirus but lack of the protein coat. • The first step in their transposition is the transcription of the entire transposon, producing an RNA copy of the element that is typically several thousand nucleotides long. This
- 16. Biology Elite biologyelite.weebly.com transcript, which is translated as a messenger RNA by the host cell, encodes a reverse transcriptase enzyme. This enzyme makes a double-strand DNA copy of the RNA molecule via an RNA–DNA hybrid intermediate, precisely mirroring the early stages of infection by a retrovirus. Then, the linear double-stranded DNA is inserted into the chromosome by intergrase. • Nonretroviral retrotransposon: distinct mechanism requires a complex of endonuclease and reverse transcriptase • A significant fraction o vertebrate chromosomes is made up of repeated DNA sequence. In human, these repeats are mostly mutated version of nonretroviral retrotransposons including LINE and SINE (long/short inter spread nuclear element) • Some of transposition will lead to human diseases, for example, L1 insertion into gene- coding blood-clotting protein factor VIII will cause haemophilia Conservative site-specific recombination: • Breaking and rejoining DNA sequence at two specific site. • Depending on the position and orientation, it can be classified into DNA integration, DNA excision and DNA inversion
- 17. Biology Elite biologyelite.weebly.com • DNA virus can use this machismo to move their genome in and out the host cell easily. • Conservative site-specific recombination can be also used in control of gene expression. • Gene inversion can change the orientation of the promoter genes and therefore change the gene expression. Due to reversibility, the gene on the both side and be switch on and off easily. Transposition Conservative site-specific recombination requires only that the transposon have a specialized sequence requires specialized DNA sequences on both the donor and recipient DNA does not proceed through a covalently joined protein–DNA intermediate recombinases that catalyze conservative site-specific recombination resemble topoisomerases in the sense that they form transient high-energy covalent bonds with the DNA and use this energy to complete the DNA rearrangements leaves gaps in the DNA that must be repaired by DNA polymerases. No gaps