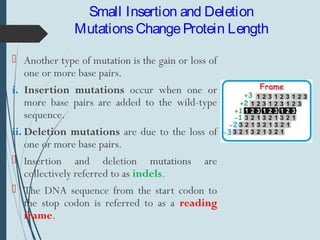
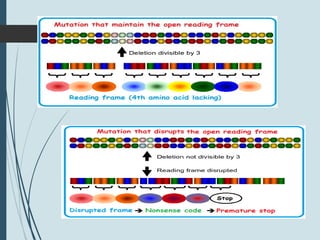
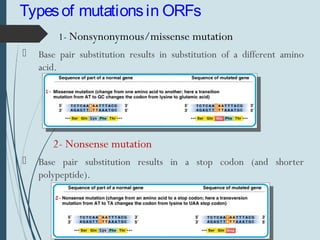
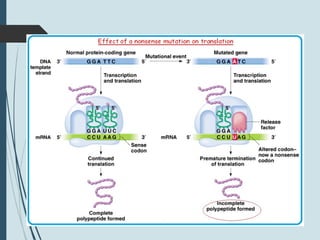
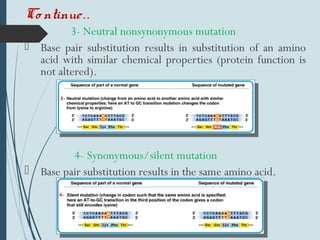
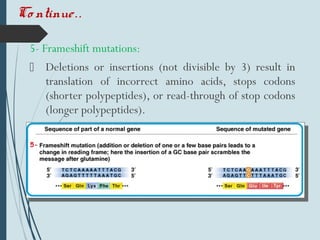
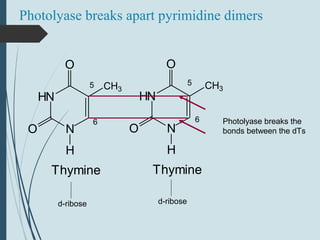
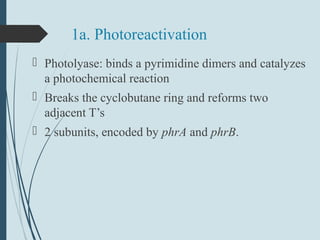
1. DNA is constantly exposed to damage from the environment and errors during replication. Cells have several DNA damage repair mechanisms to fix alterations to maintain genome integrity.
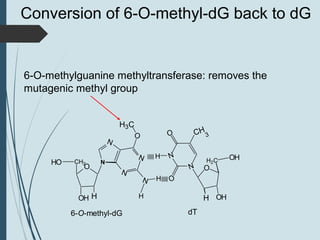
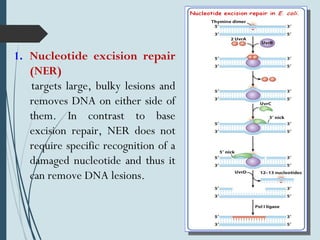
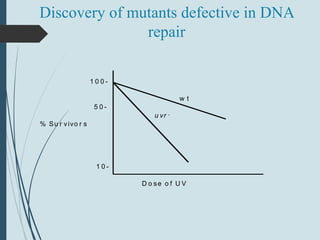
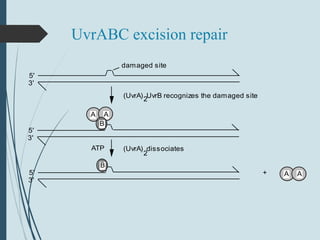
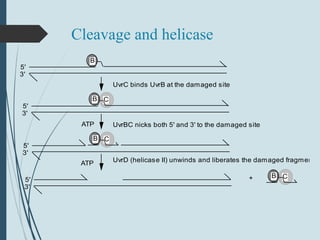
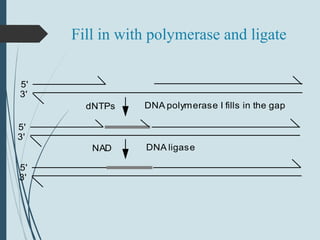
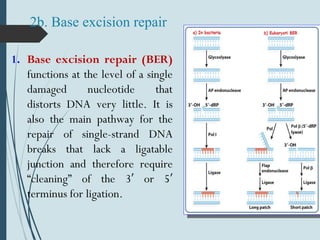
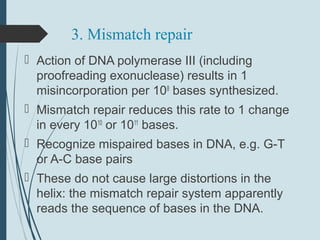
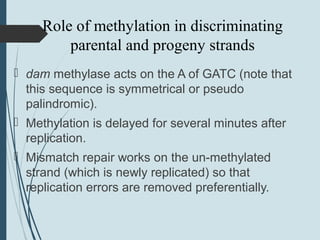
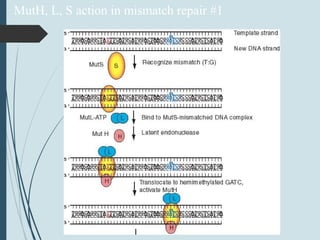
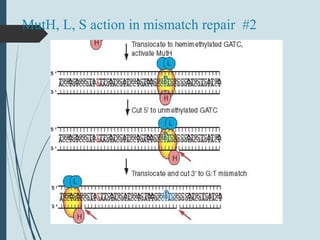
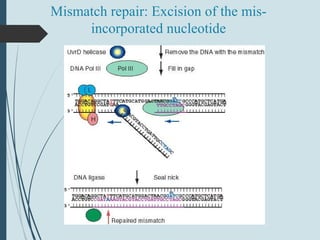
2. The main repair pathways are direct reversal, excision repair including nucleotide excision repair and base excision repair, and mismatch repair which fixes errors made during replication.
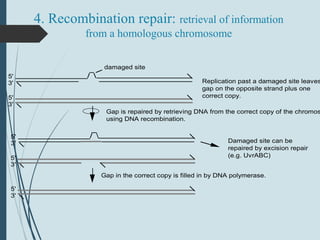
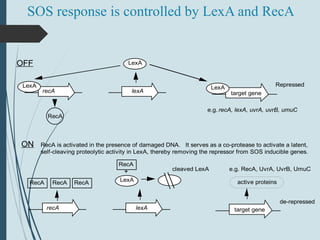
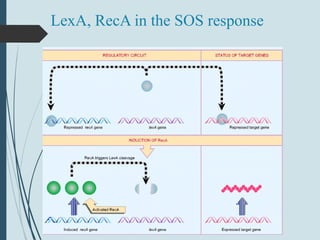
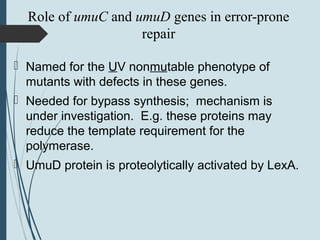
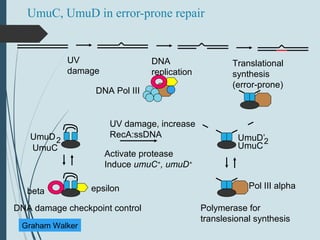
3. If damage evades these pathways, error-prone translesion synthesis can occur which often introduces mutations, acting as a last resort to allow replication past lesions.