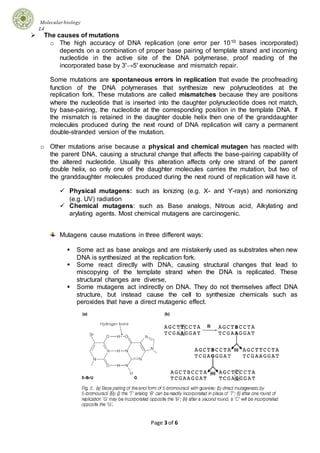
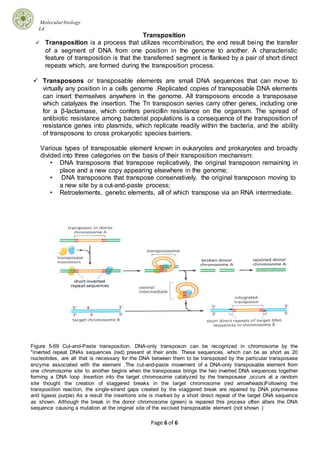
Mutations can occur through various mechanisms and can have different effects on genes and organisms. There are several types of mutations including point mutations, which alter single nucleotide pairs, and frameshift mutations, which change the reading frame. Mutations can be caused by errors in DNA replication or damage from physical or chemical mutagens. Cells have multiple DNA repair systems to correct mutations, including direct repair, excision repair, and mismatch repair. Recombination and transposition are processes that involve rearrangement of DNA sequences and can result in transfer of DNA segments within and between genomes.