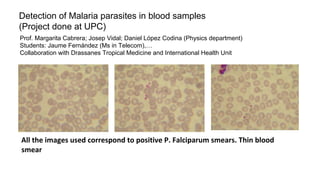
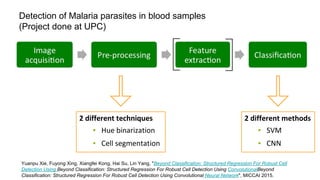
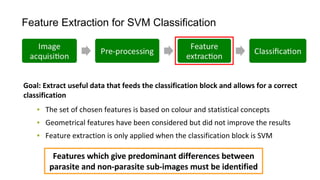
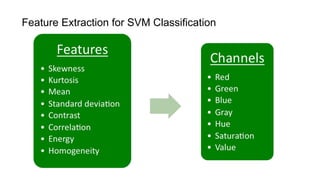
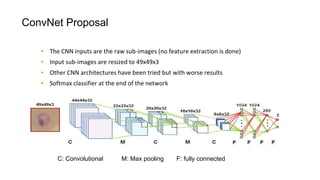
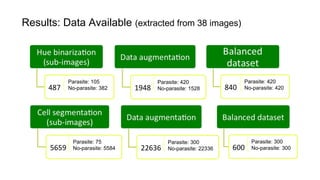
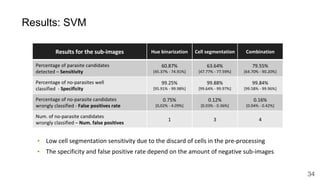
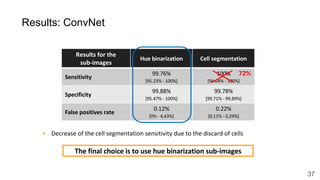
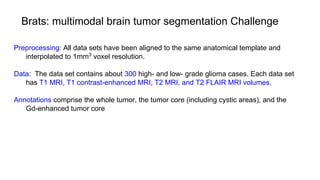
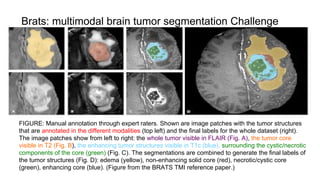
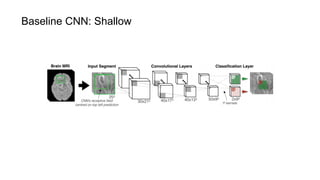
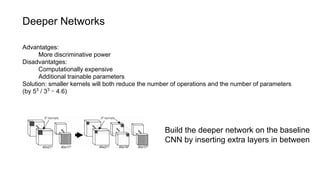
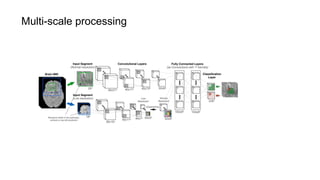
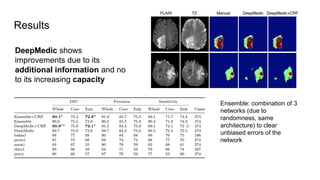
The document discusses the applications of deep learning in medical imaging, including techniques for image description, diagnosis, reconstruction, and model selection. It highlights several projects such as malaria parasite detection and brain tumor segmentation challenges, detailing methodologies and results. Key contributions from various professors and students are noted, showcasing collaborative efforts and advancements in medical image analysis.
![Day 4 Lecture 5
Medical Imaging
Elisa Sayrol
elisa.sayrol@upc.edu
[course site]](https://image.slidesharecdn.com/dlcvd4l5medicalv1-160803172703/75/Deep-Learning-for-Computer-Vision-Medical-Imaging-UPC-2016-1-2048.jpg)