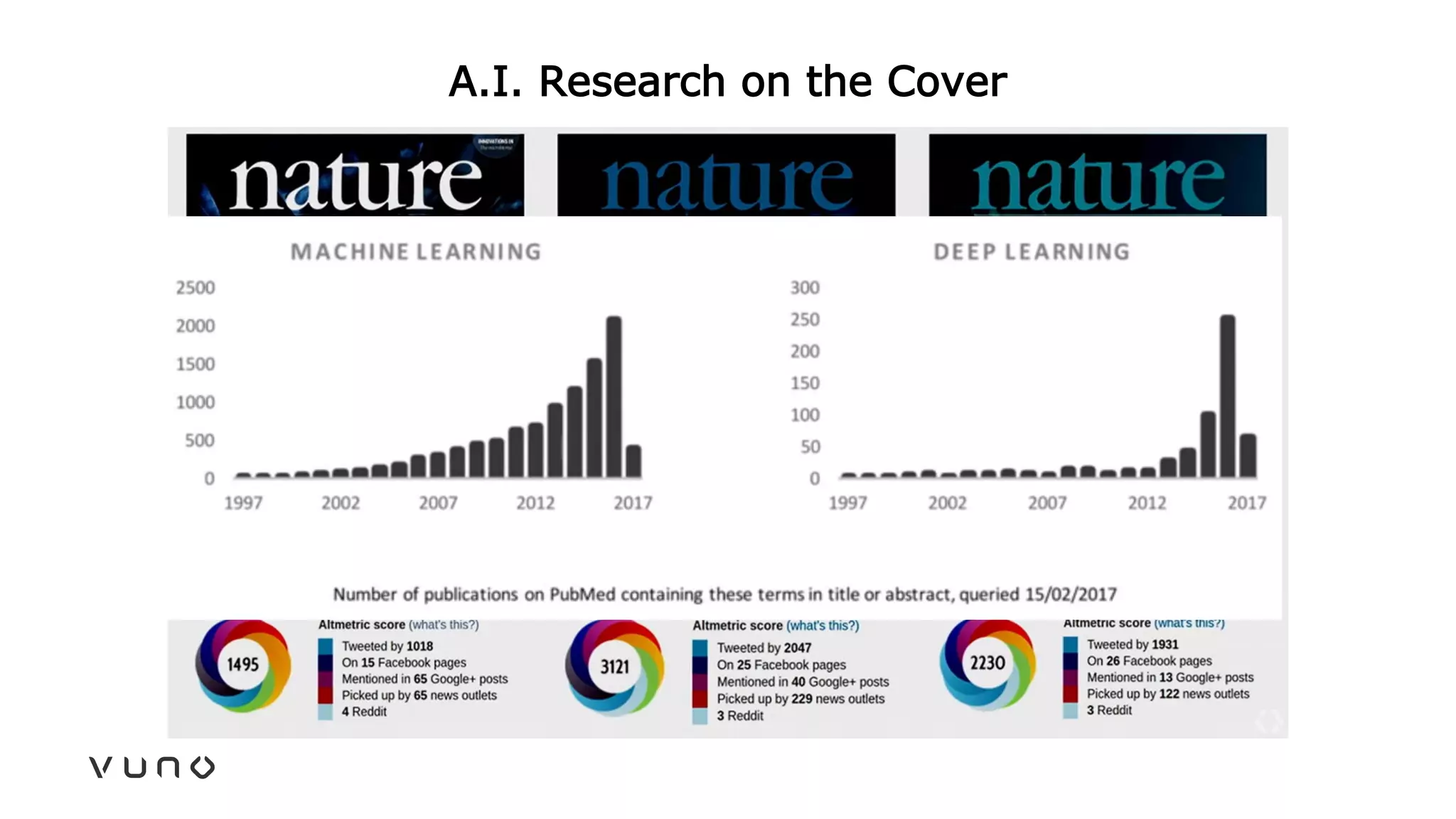
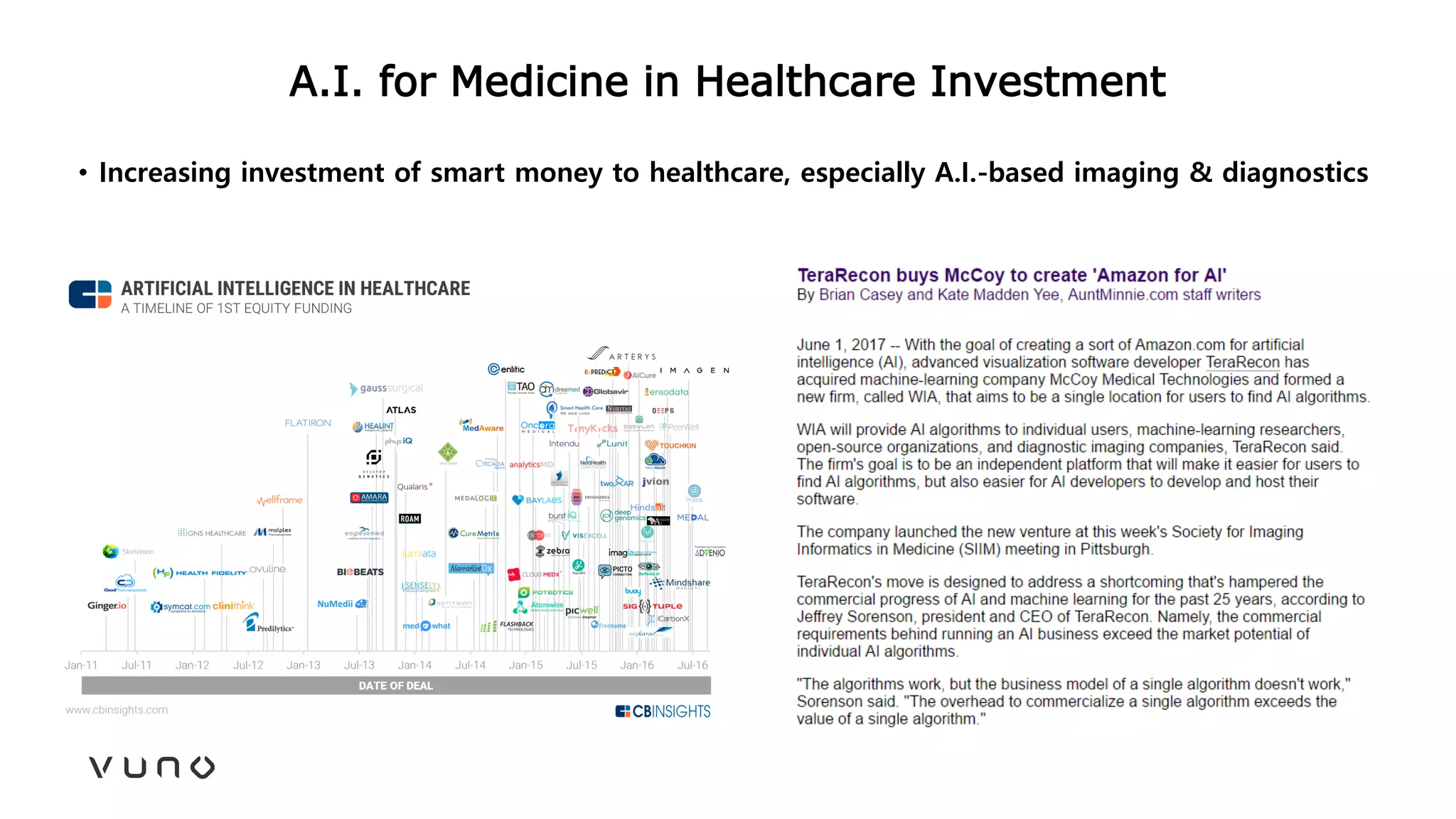
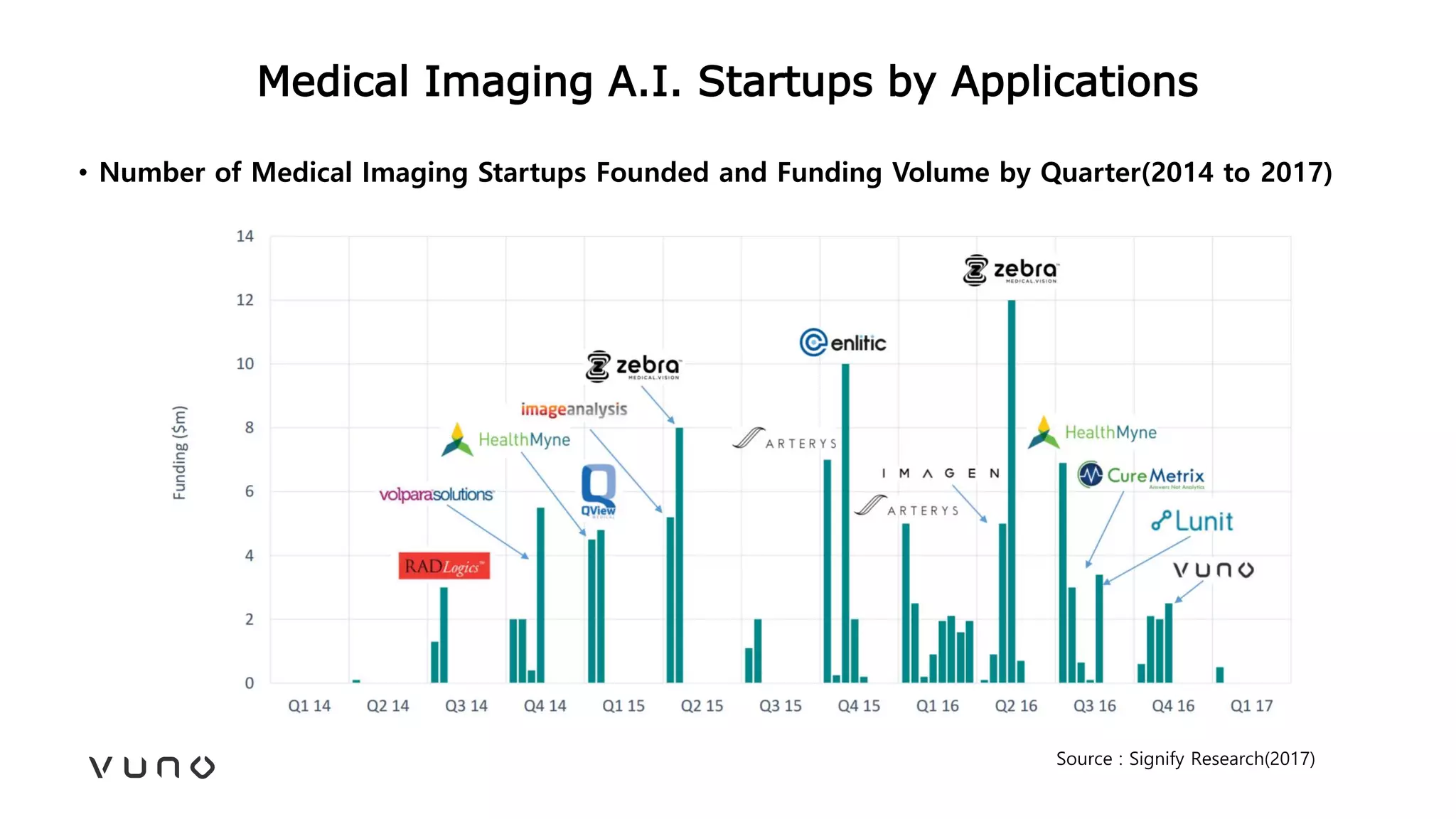
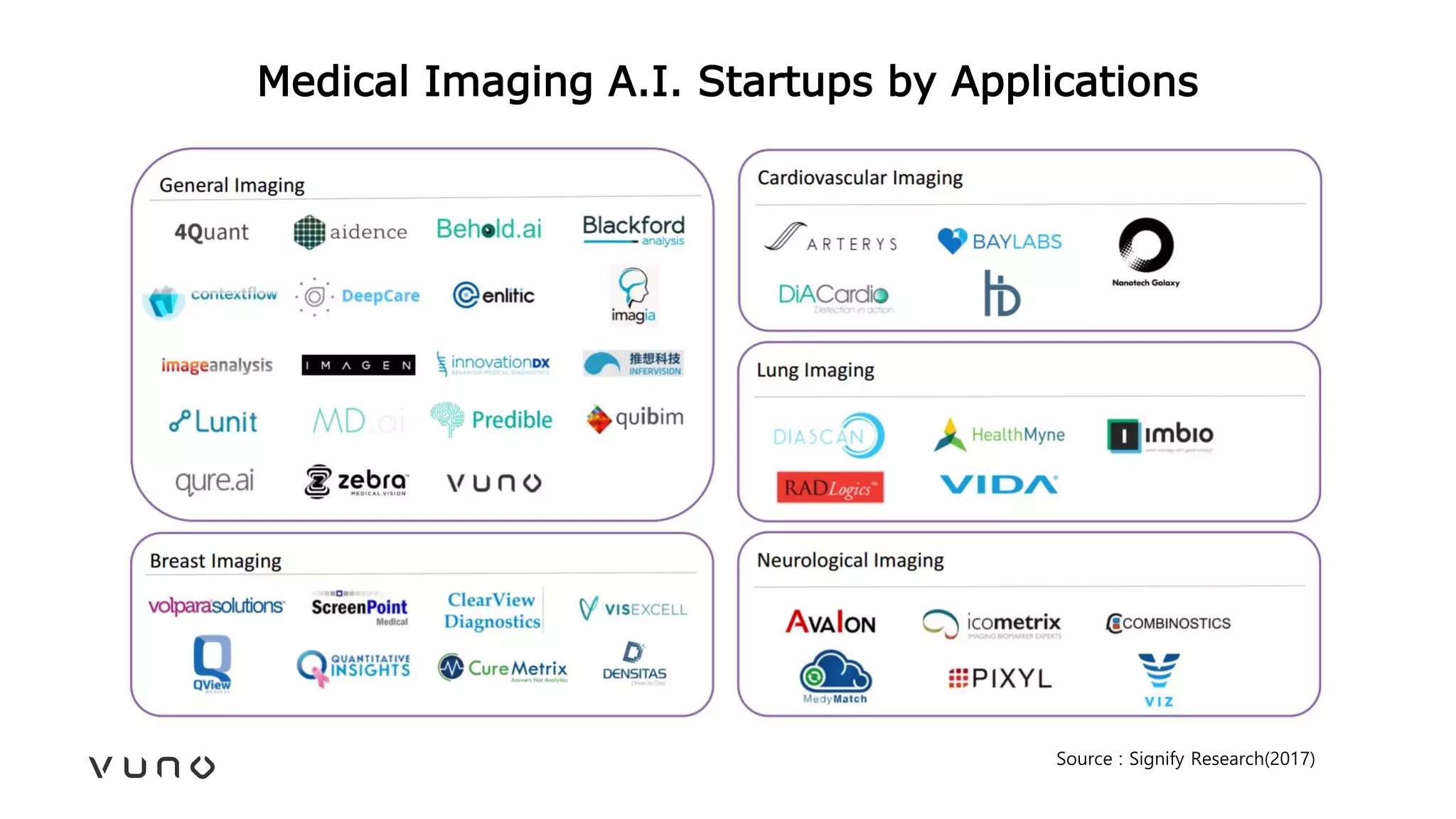
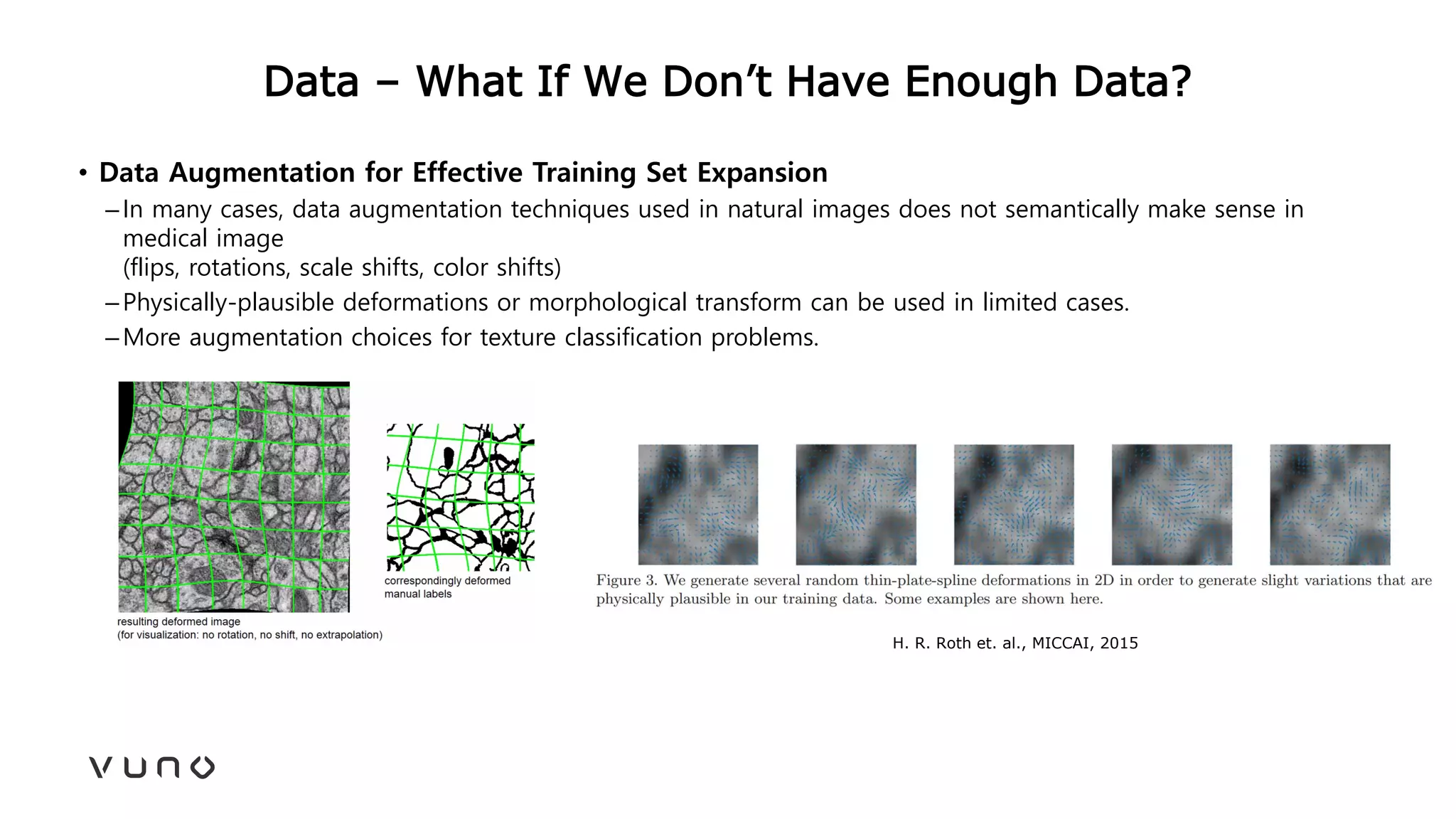
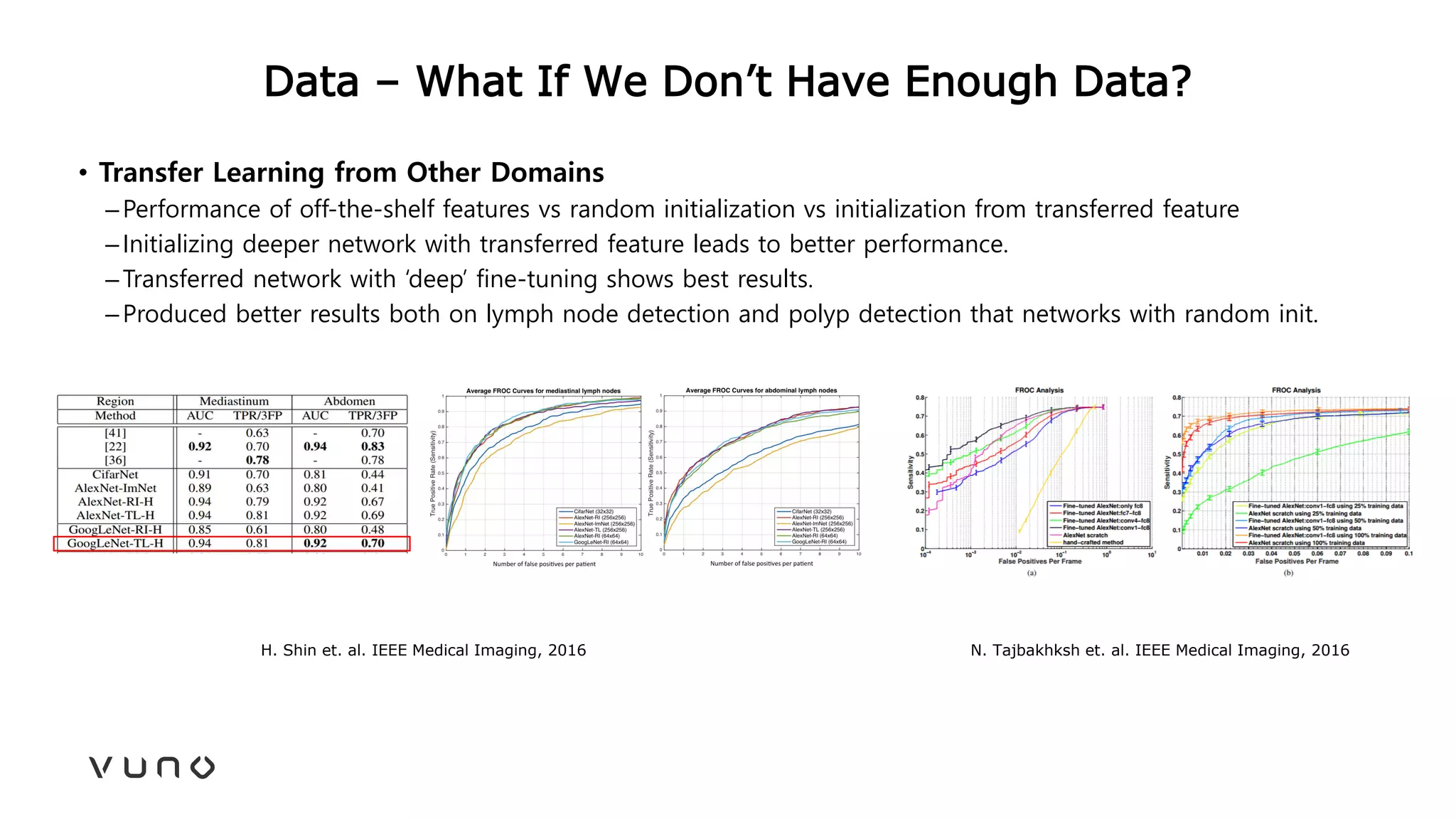
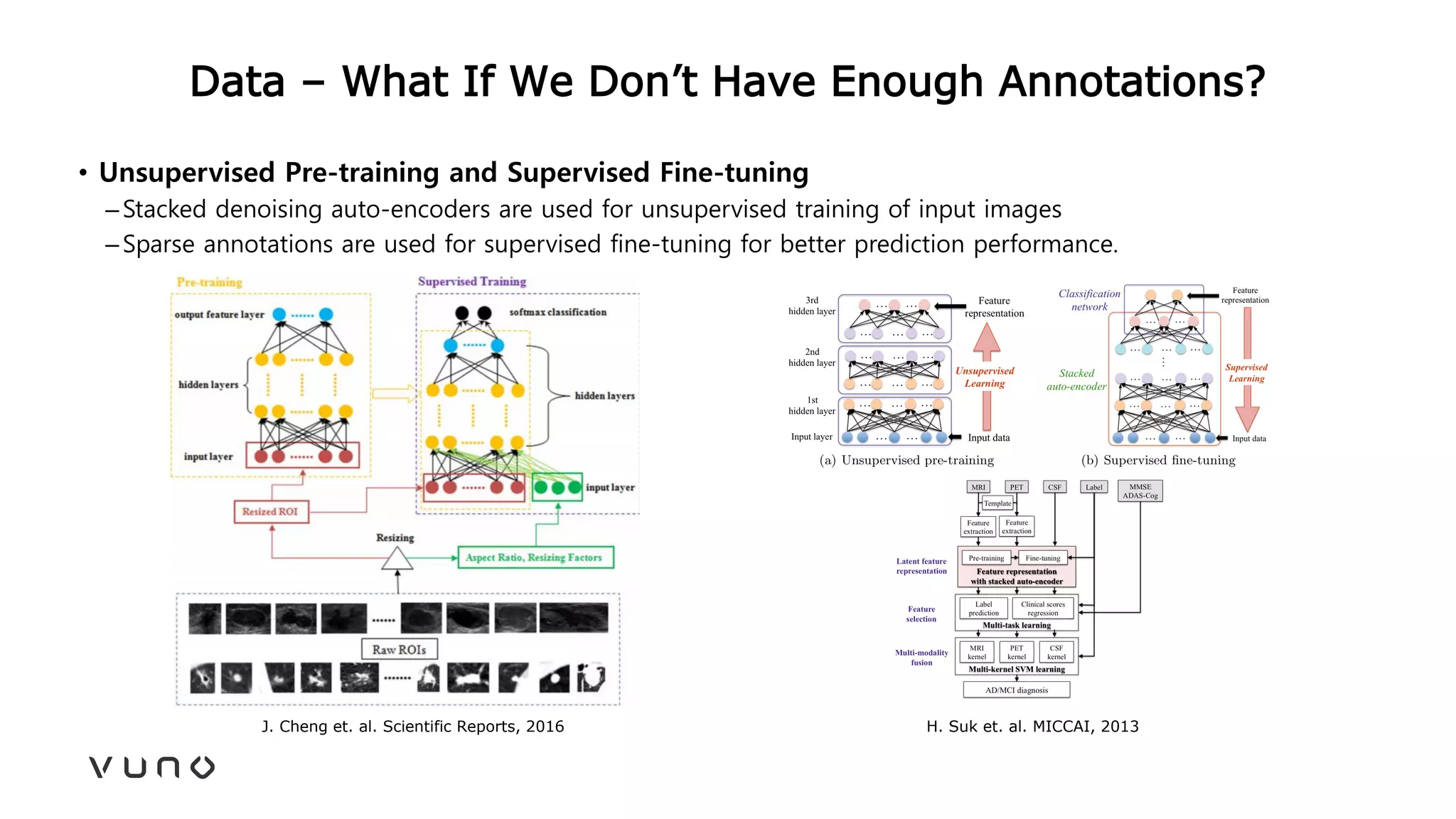
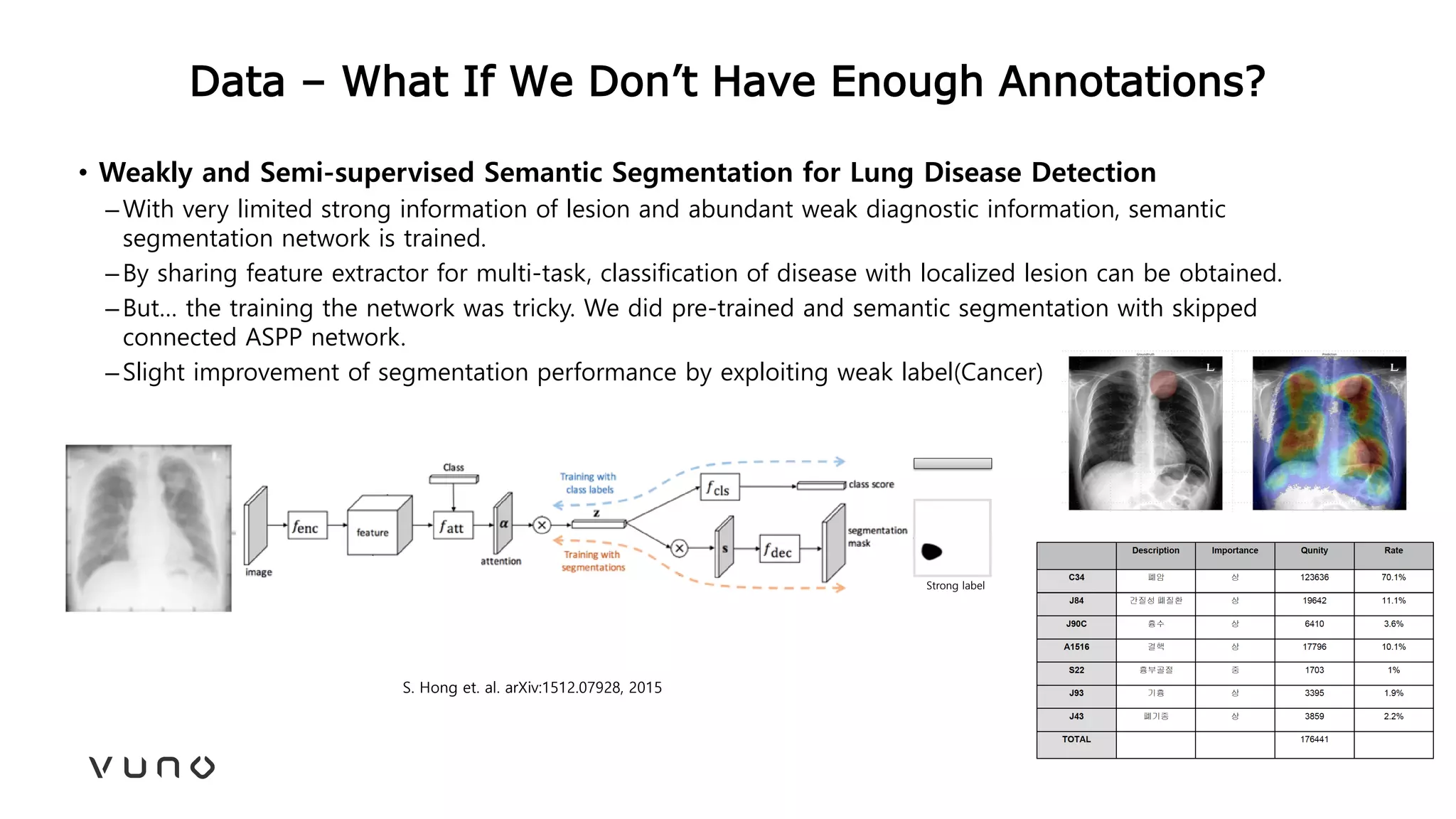
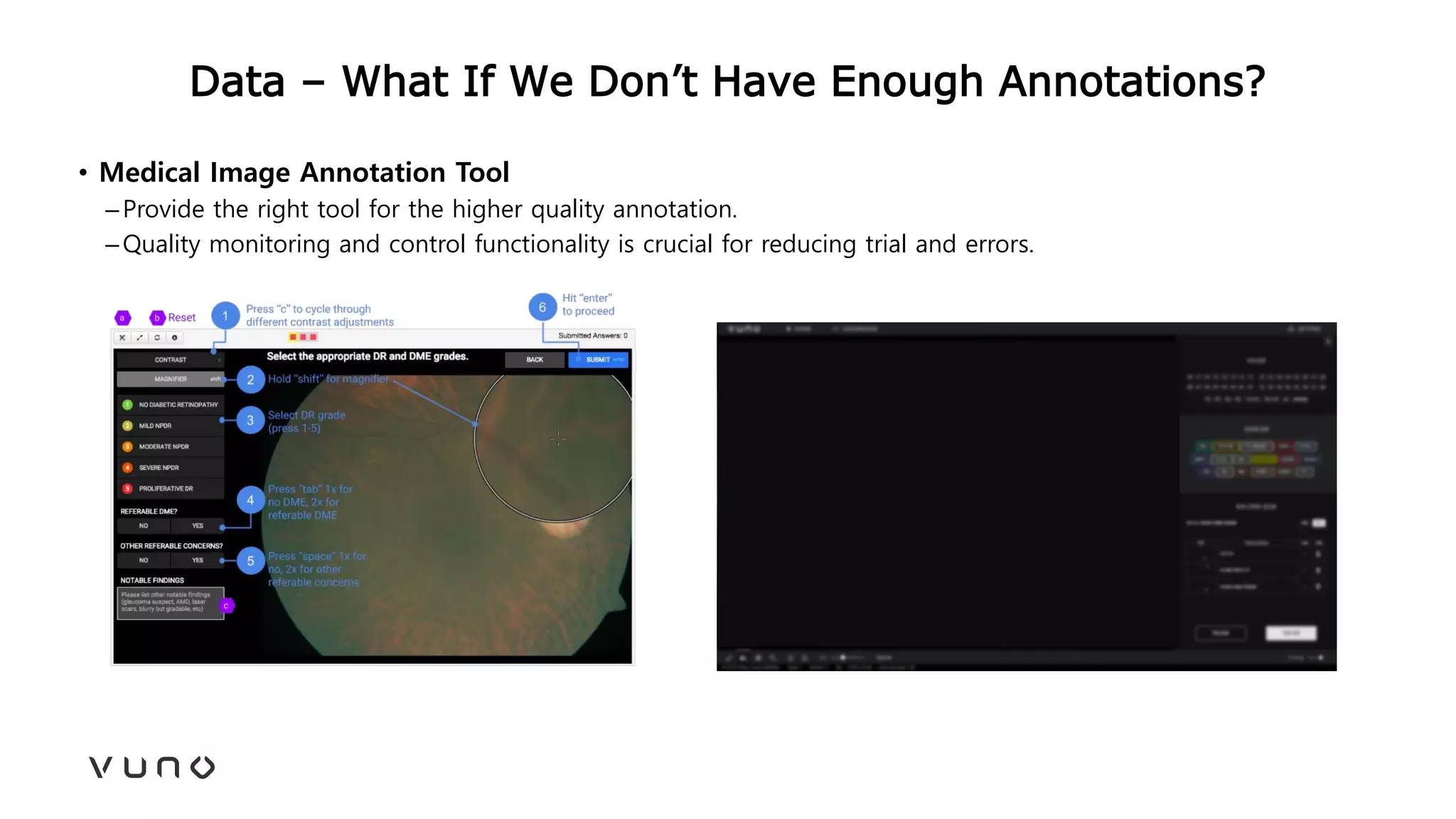
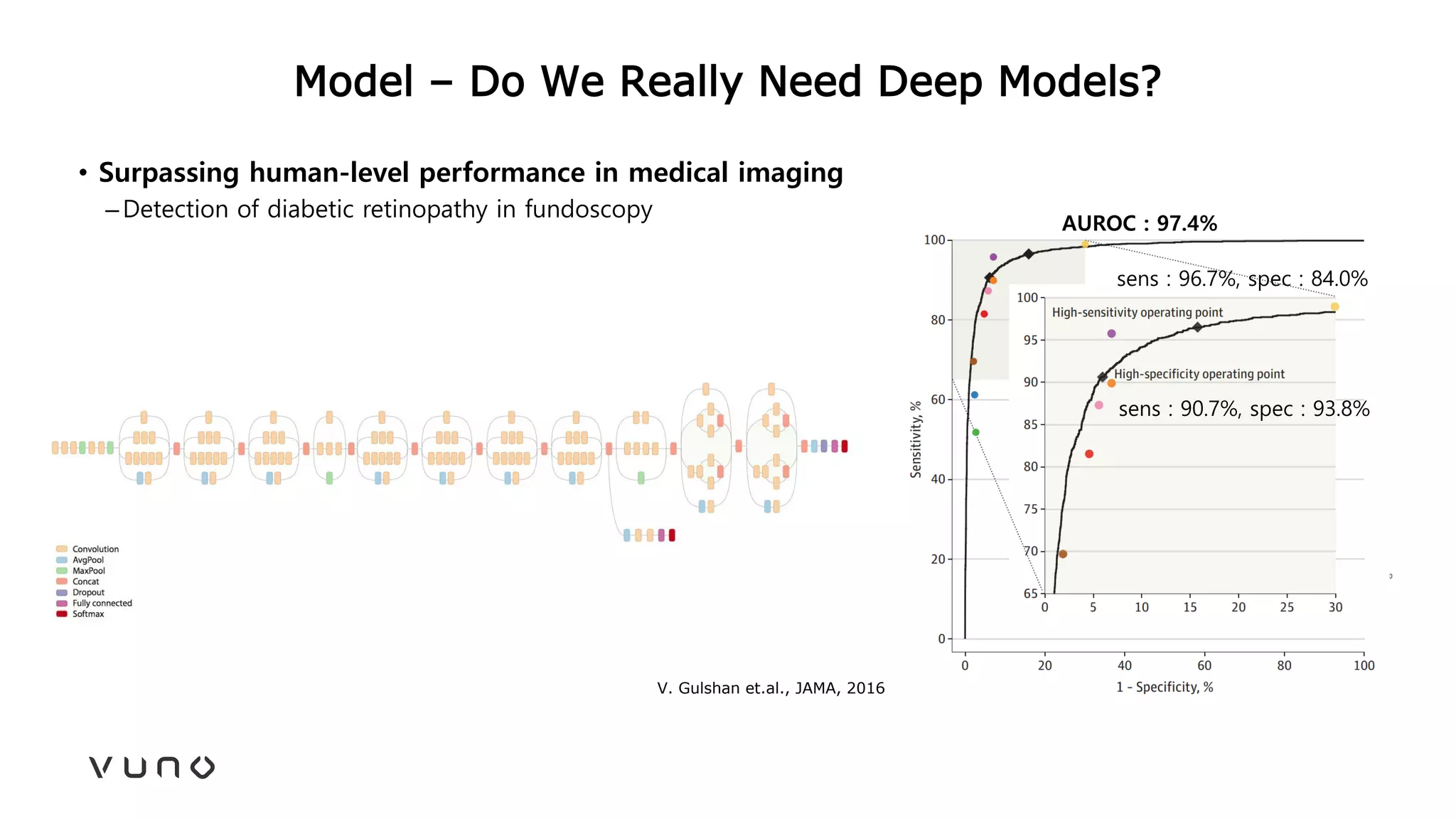
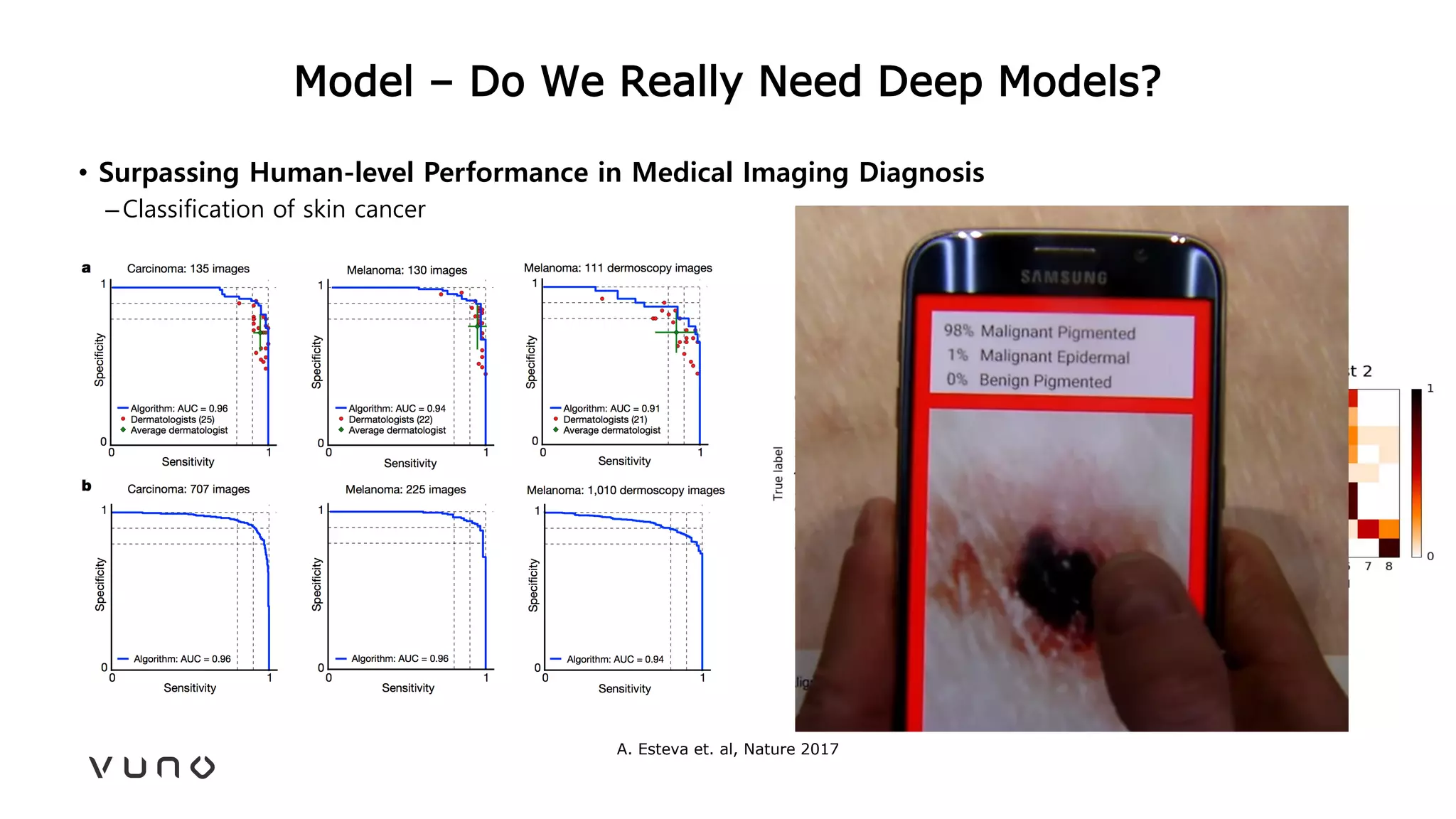
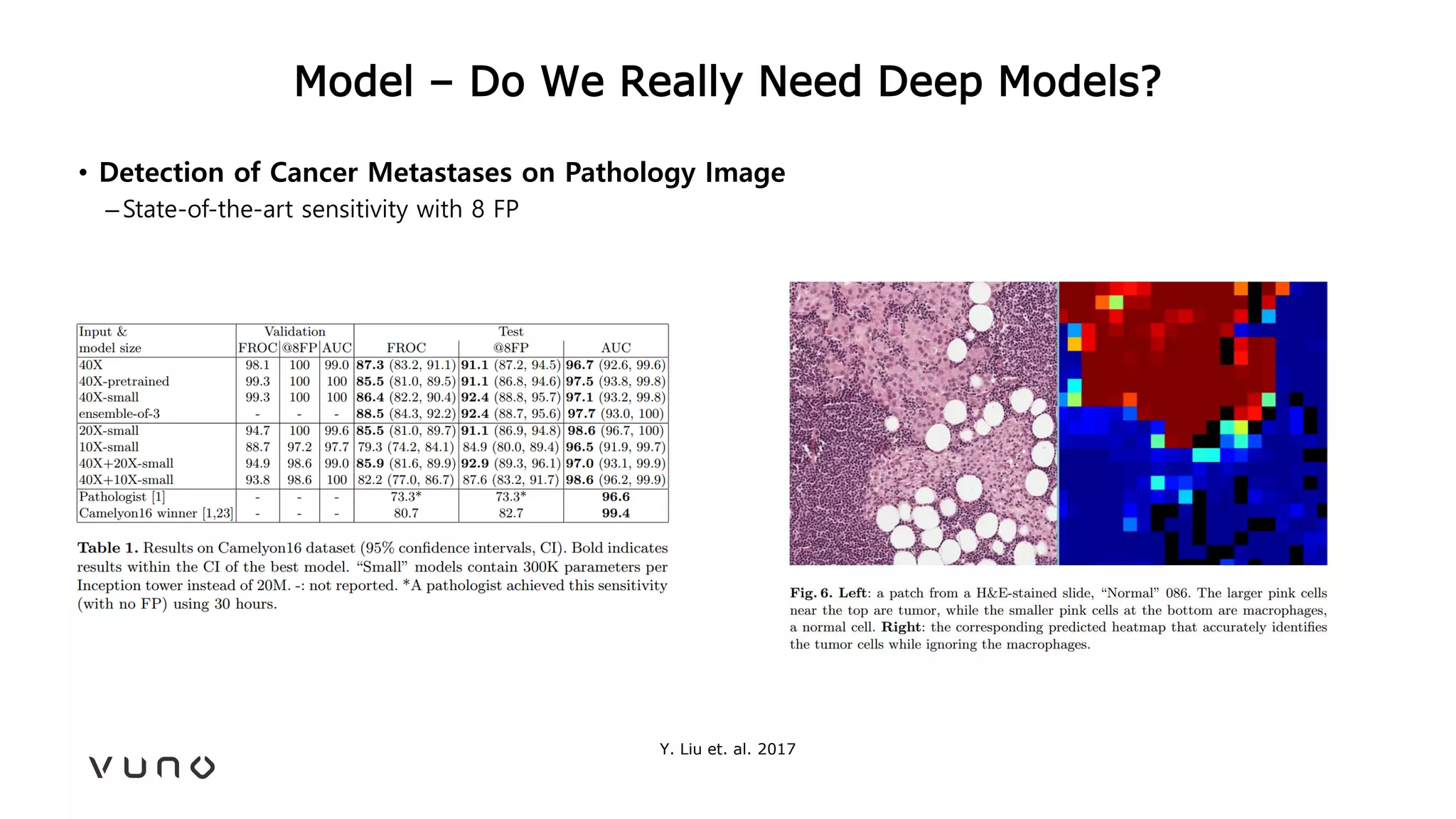
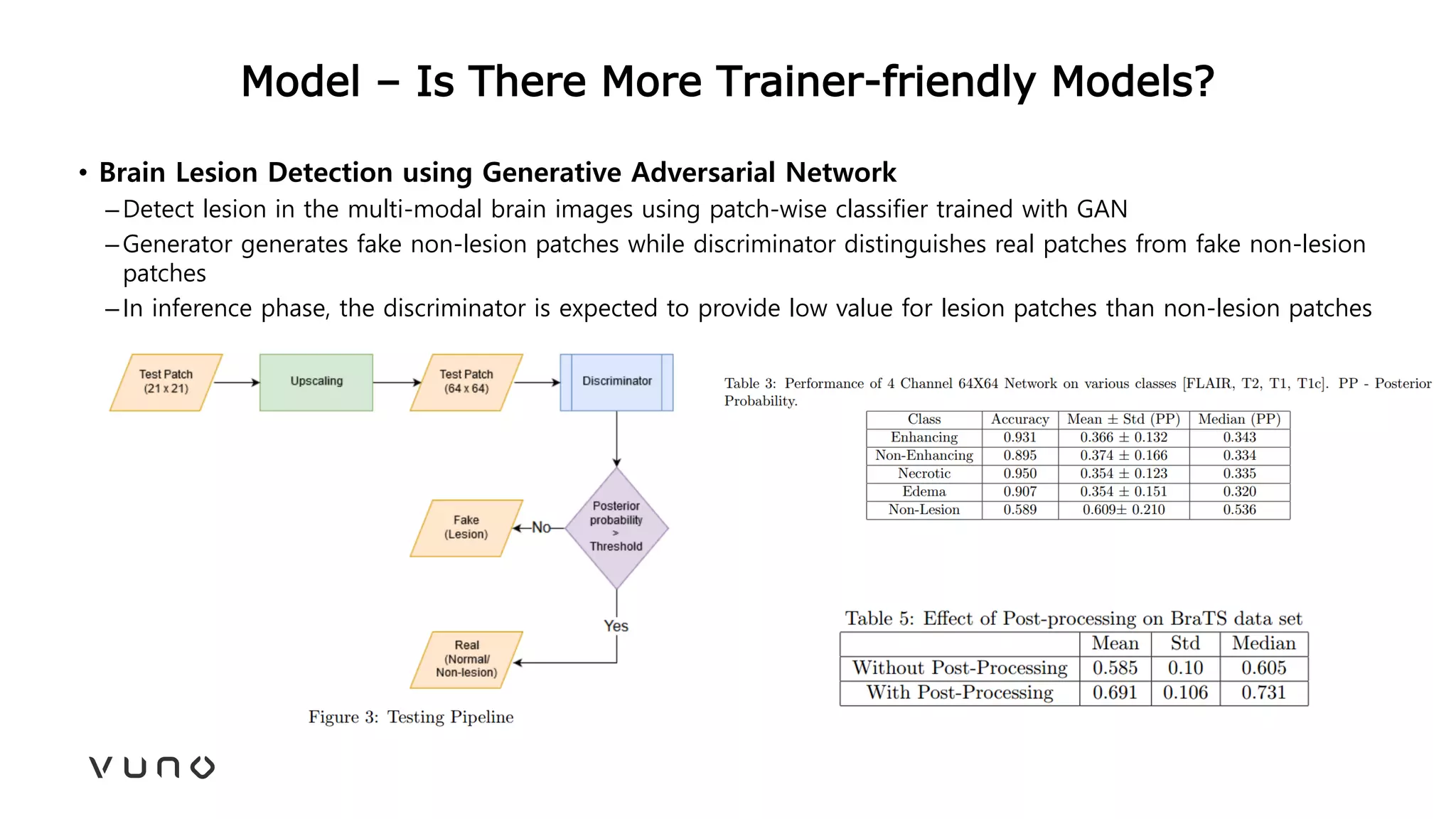
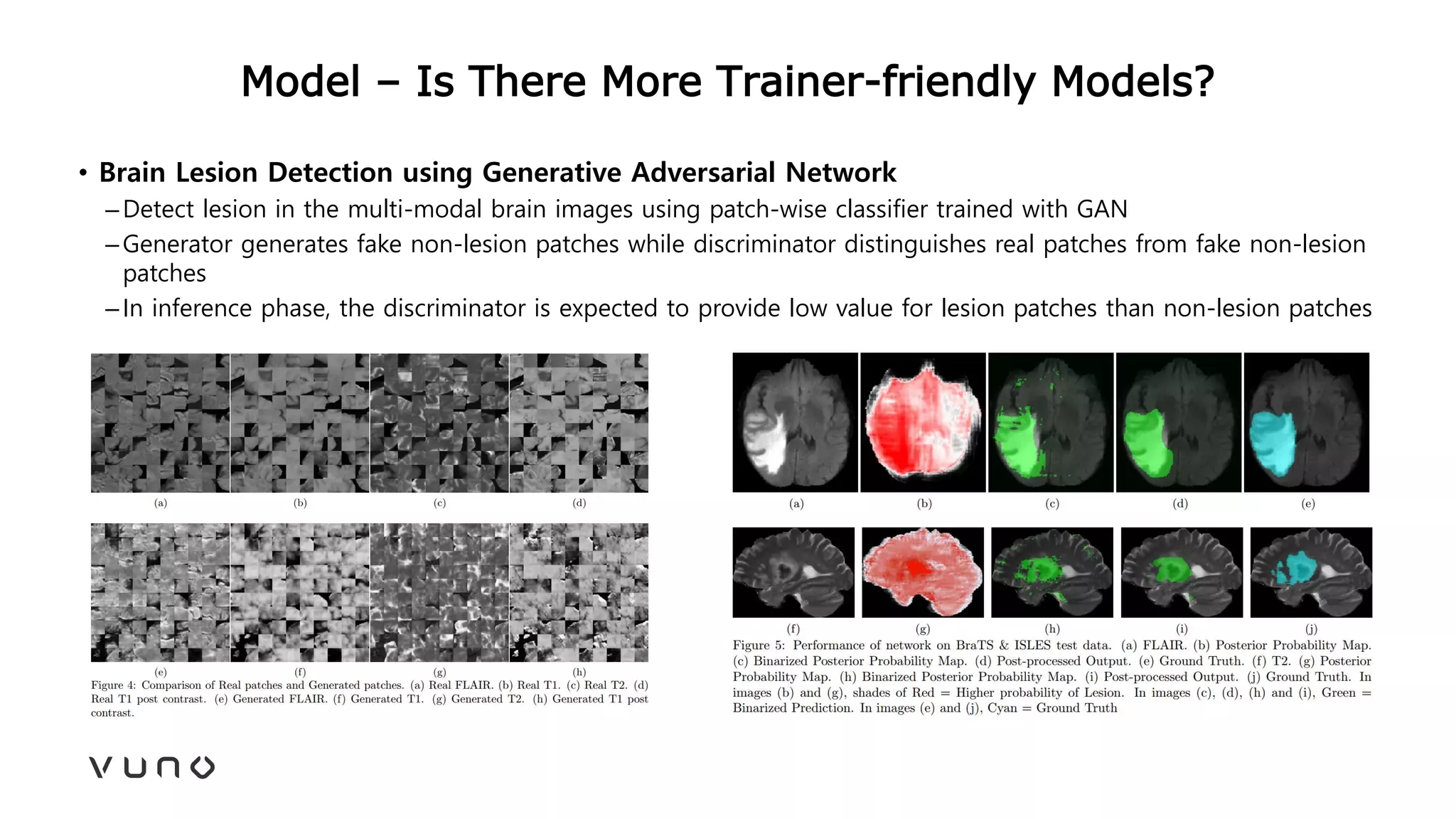
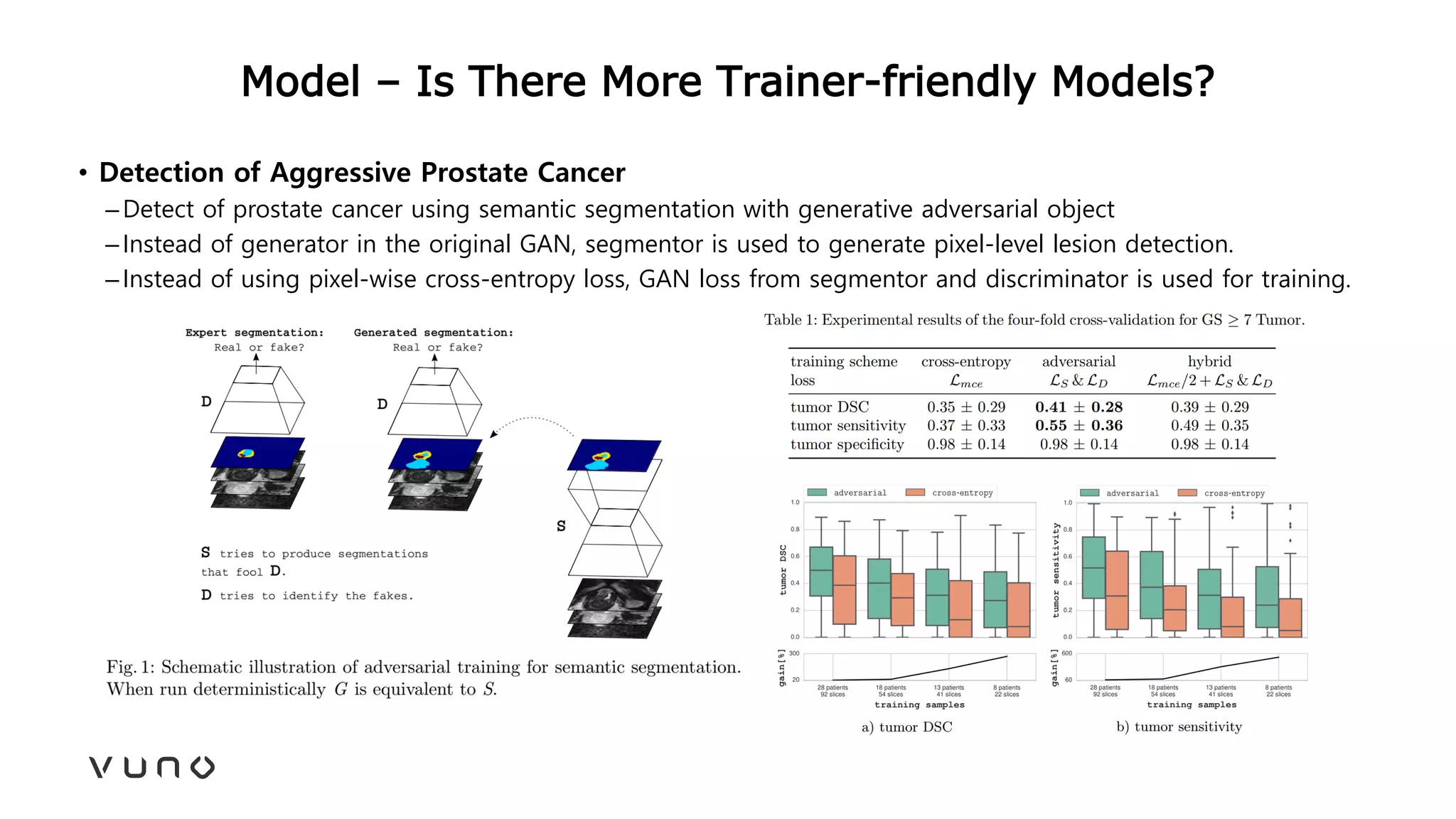
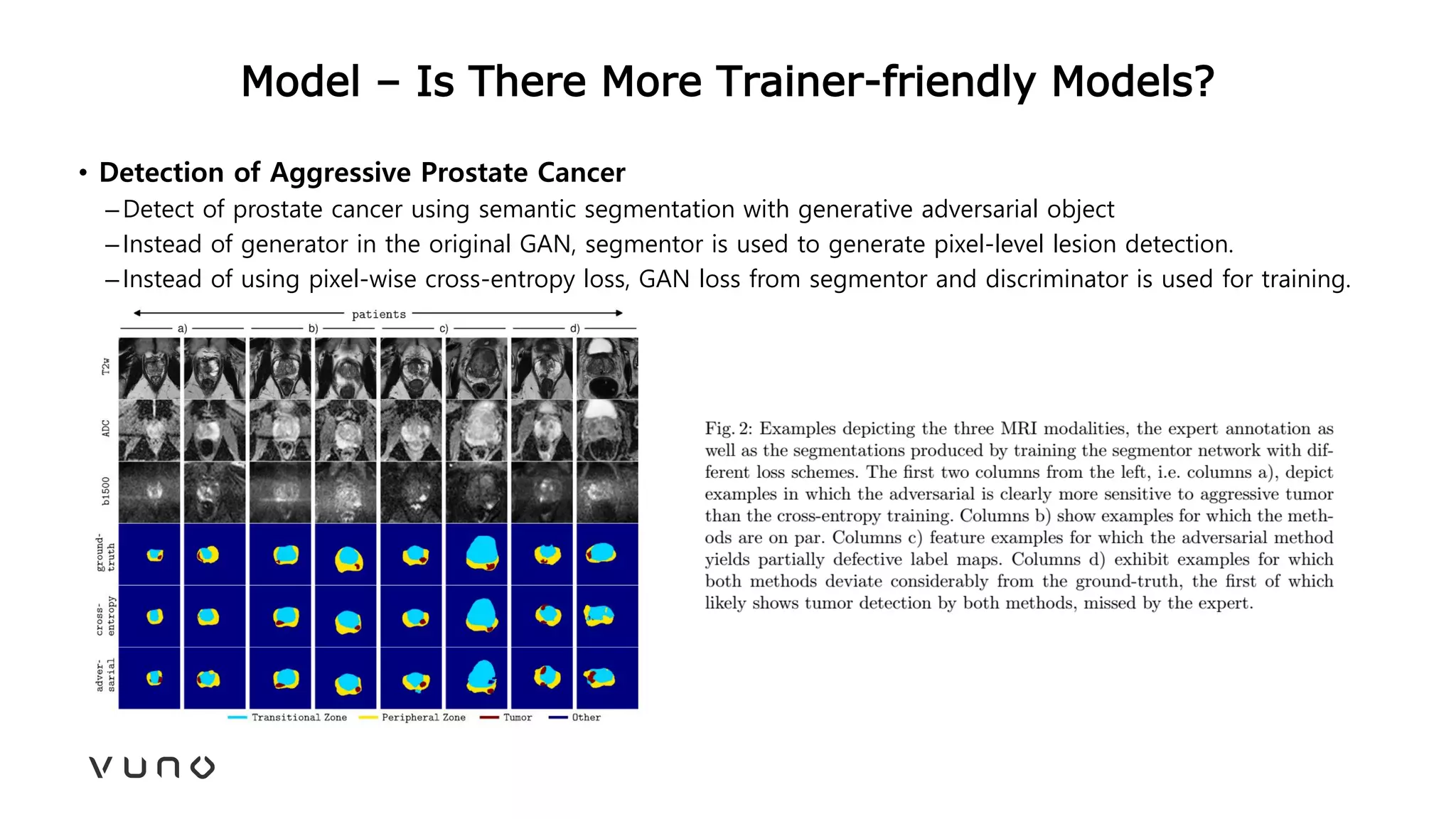
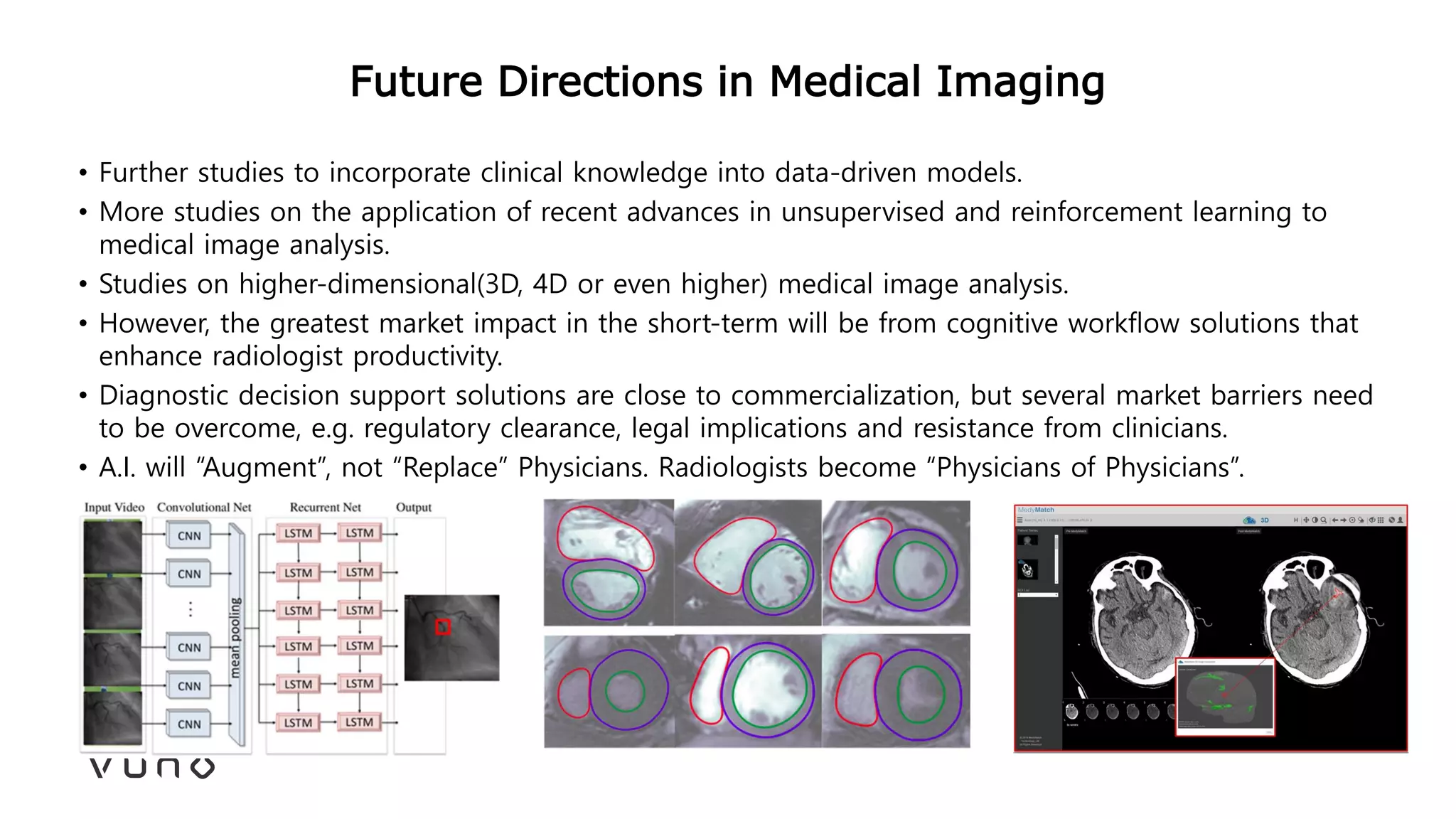
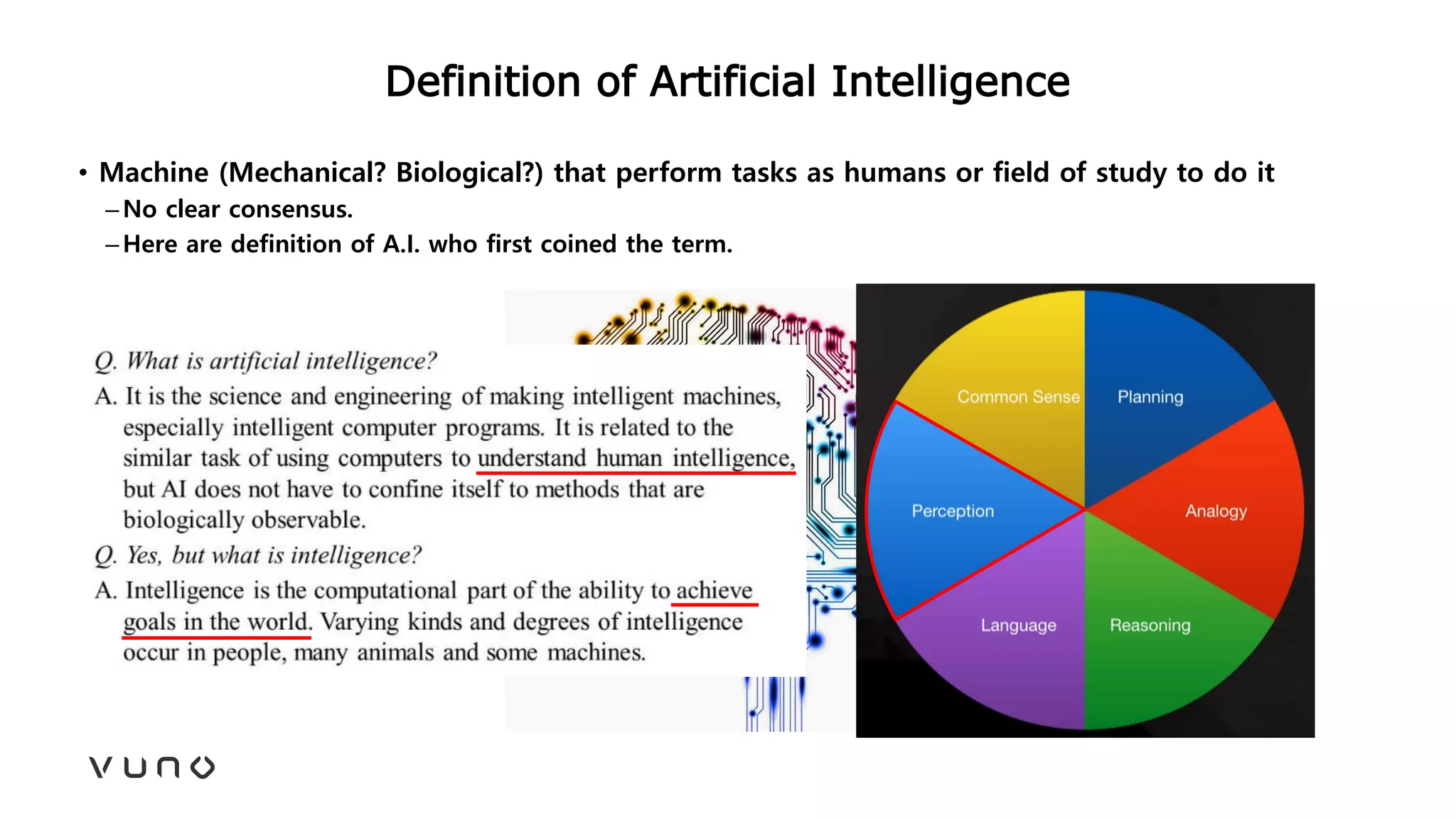
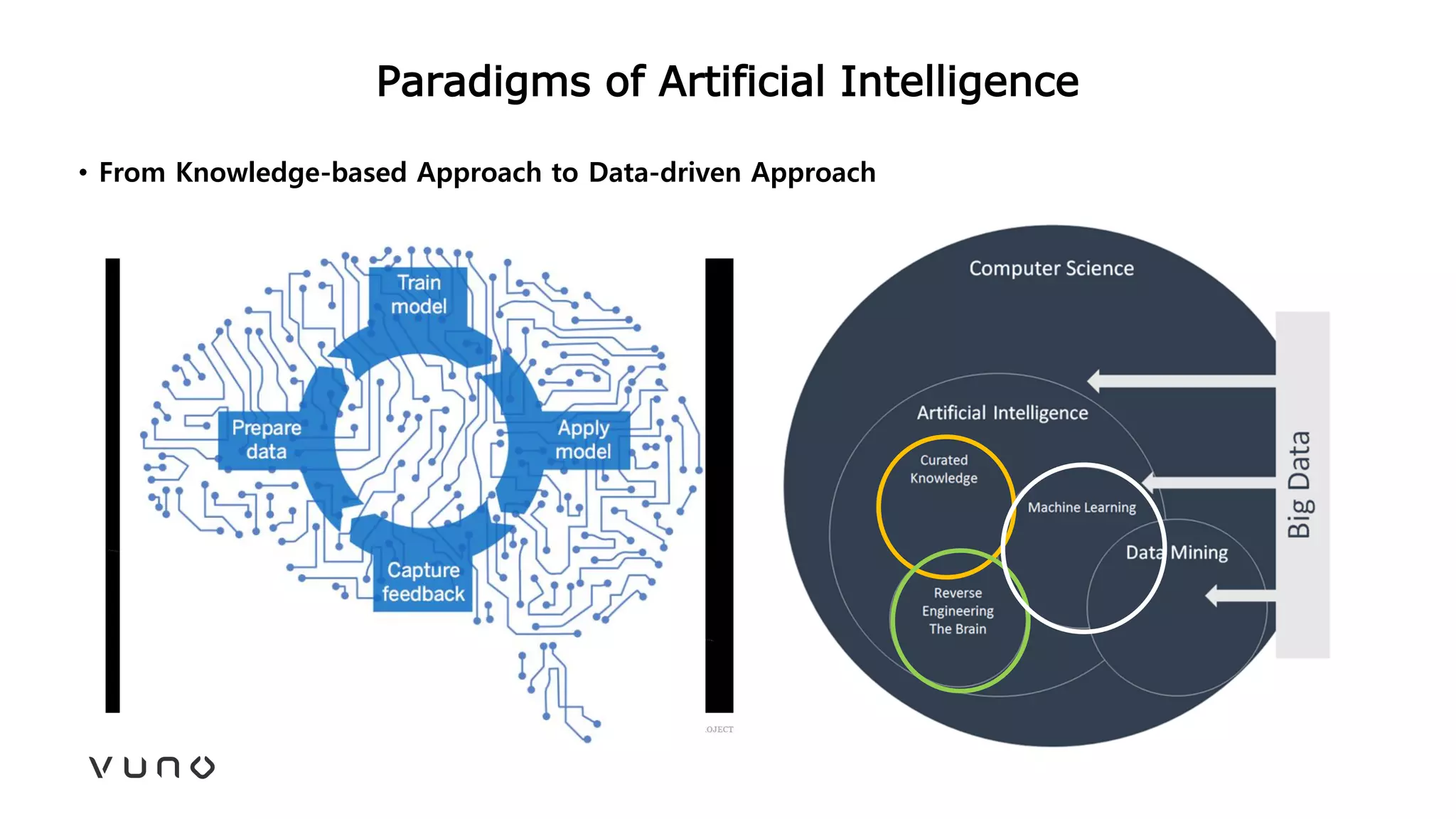
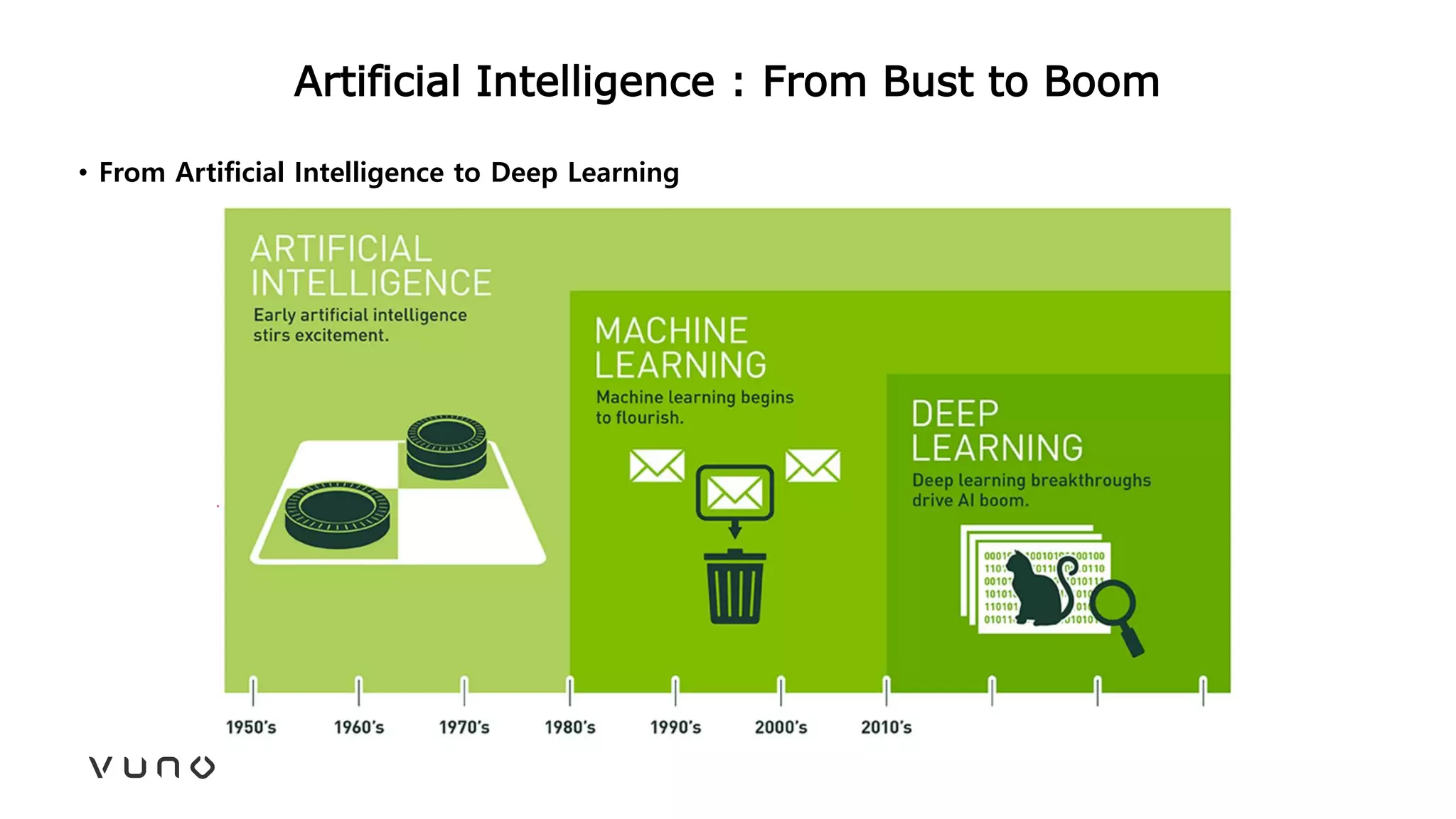
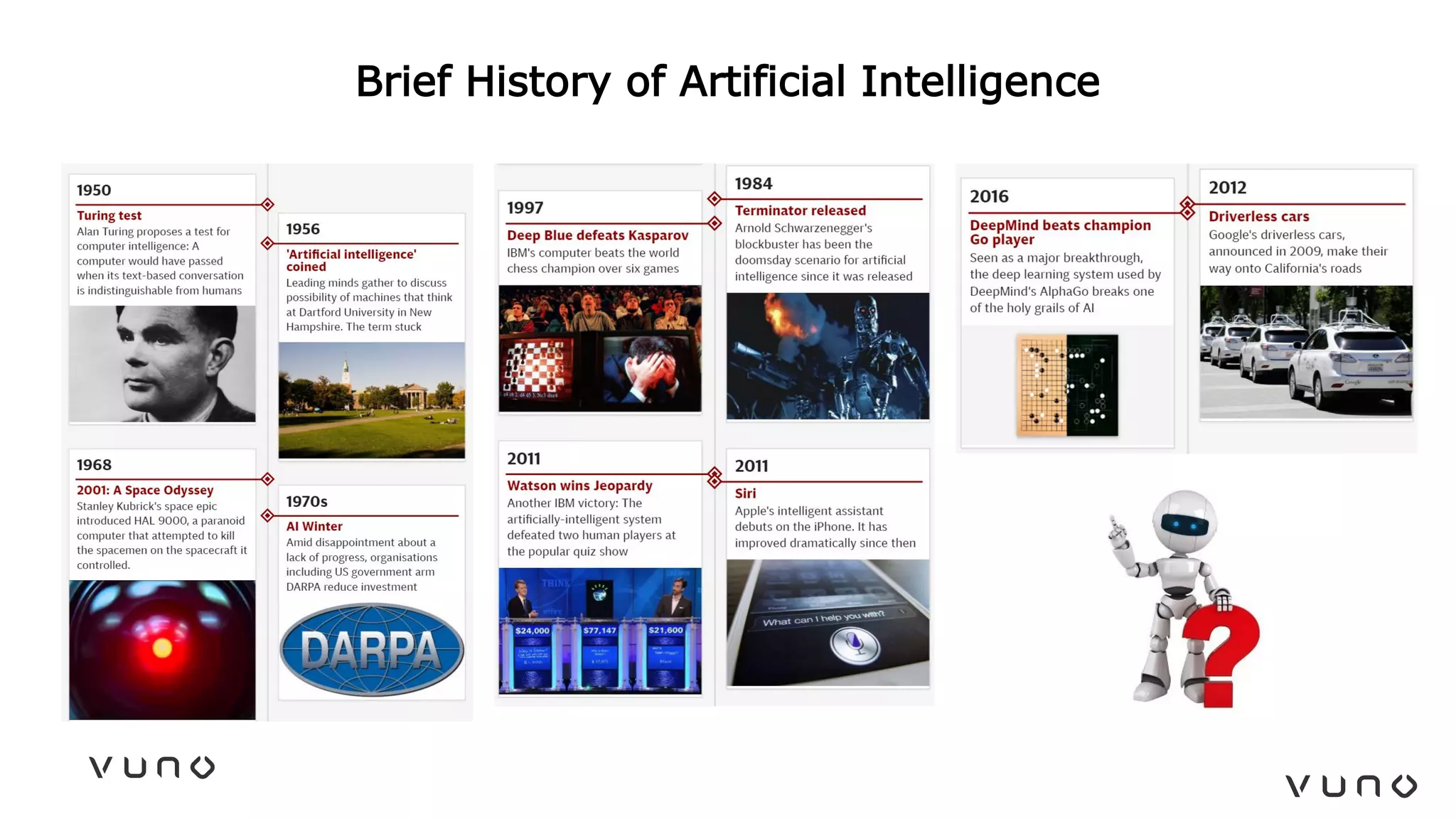
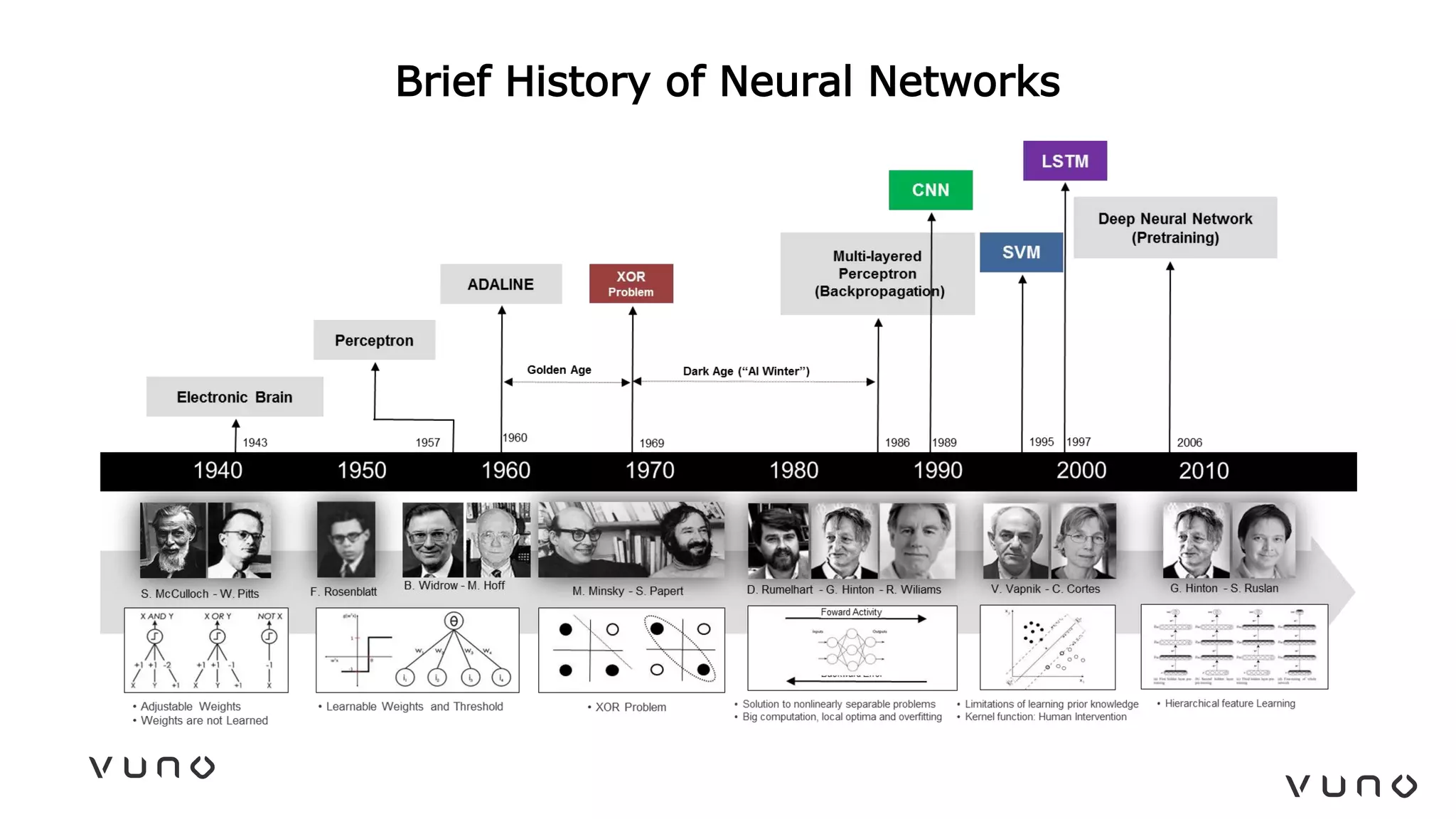
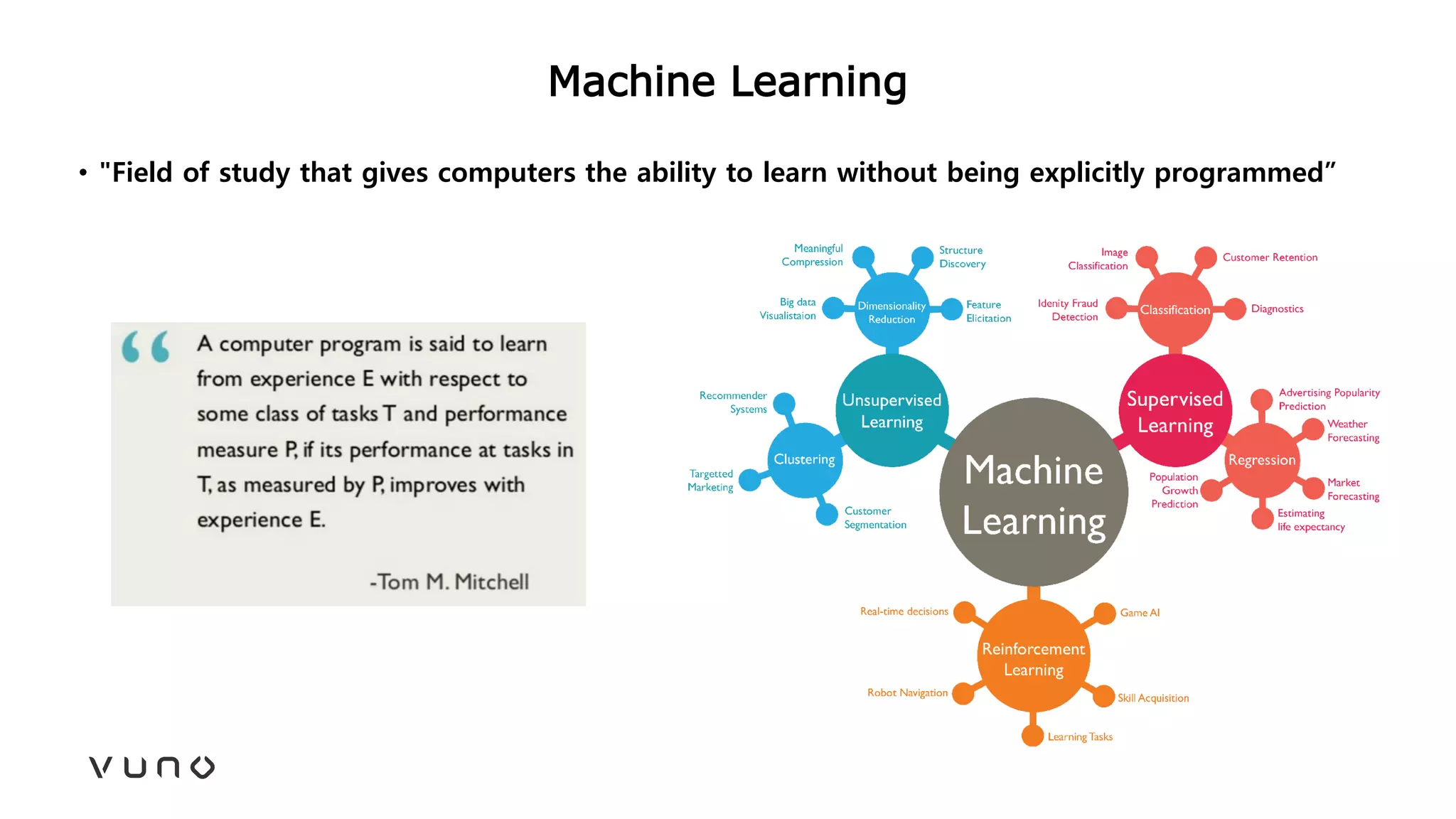
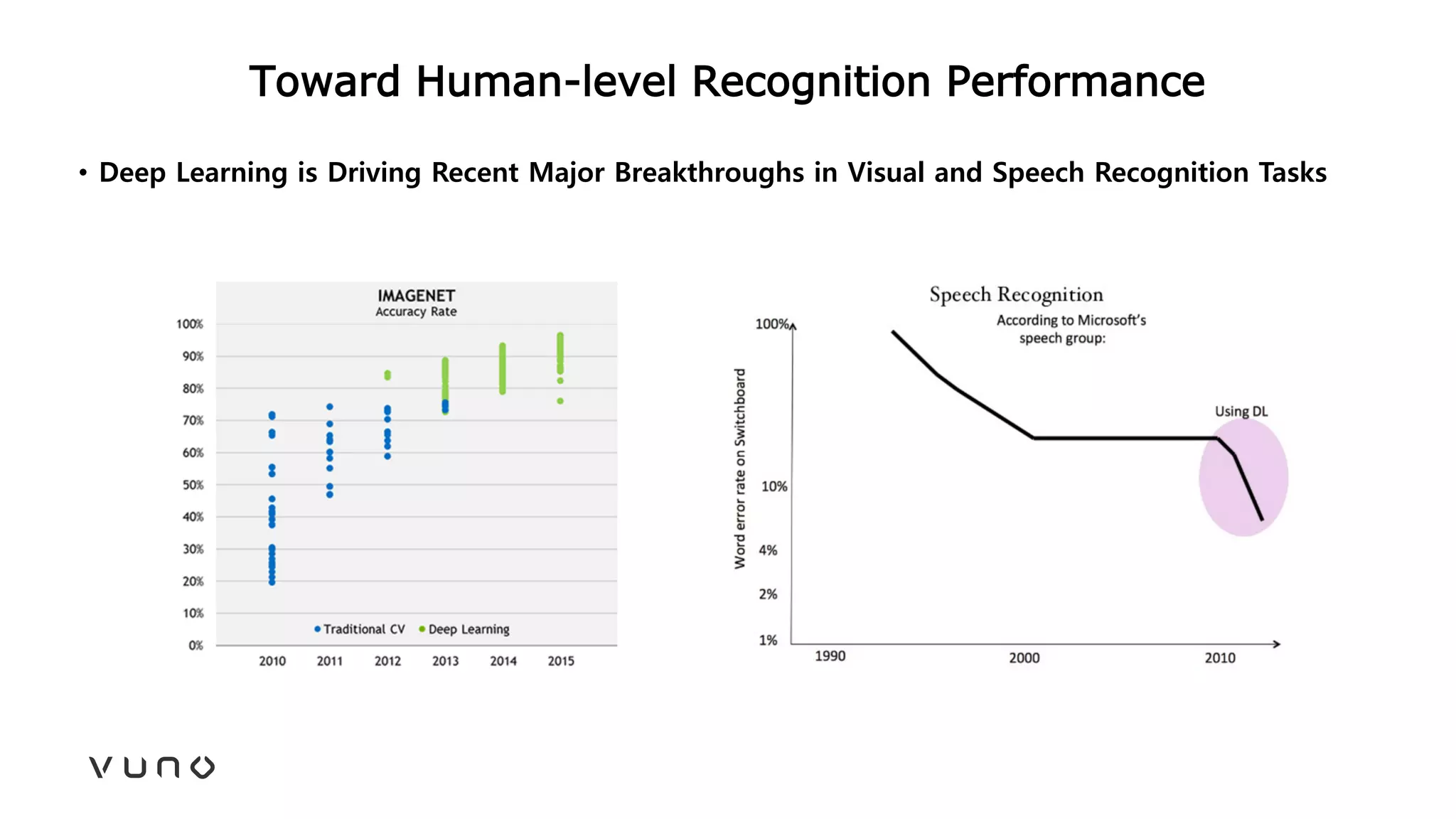
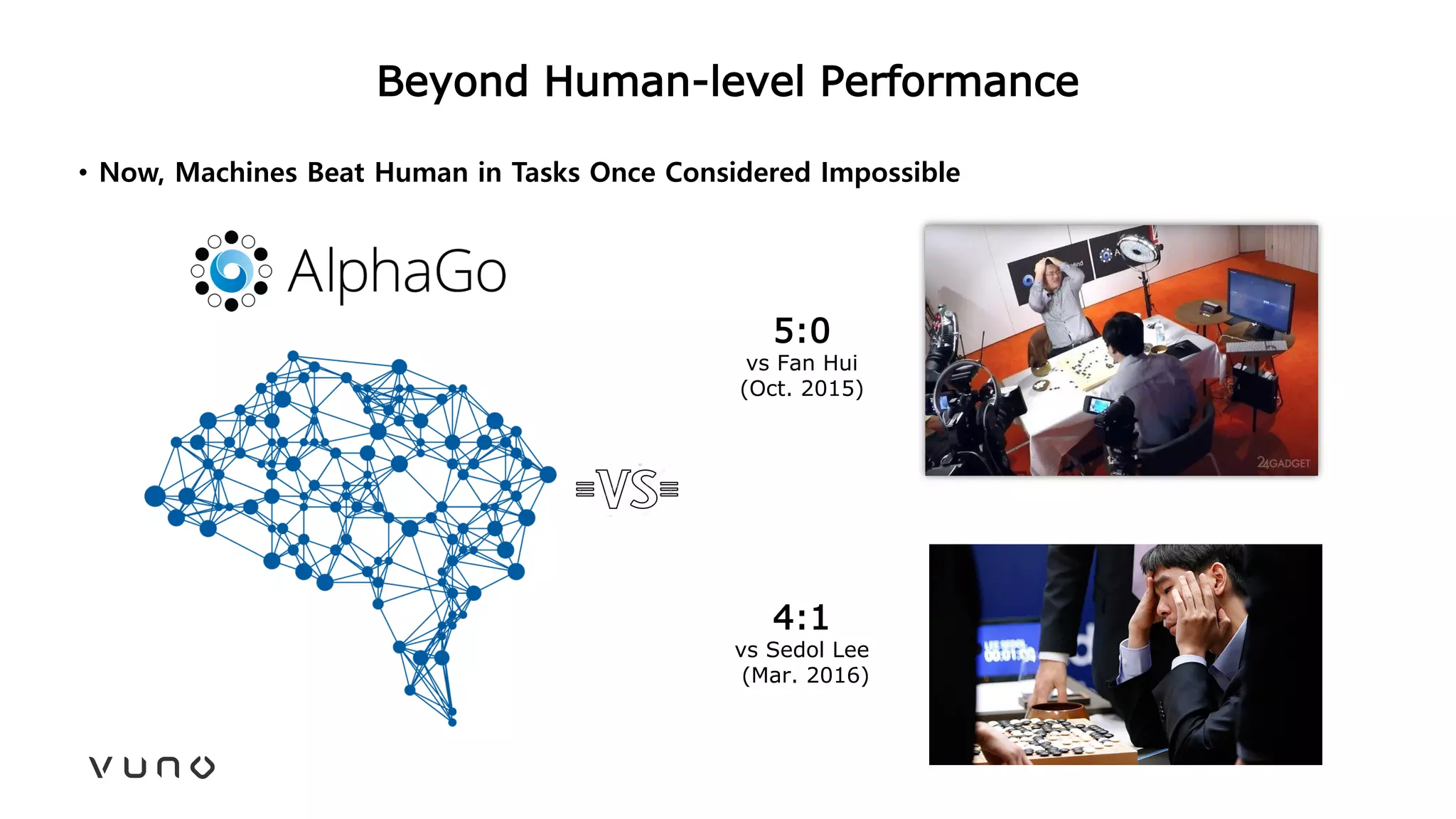
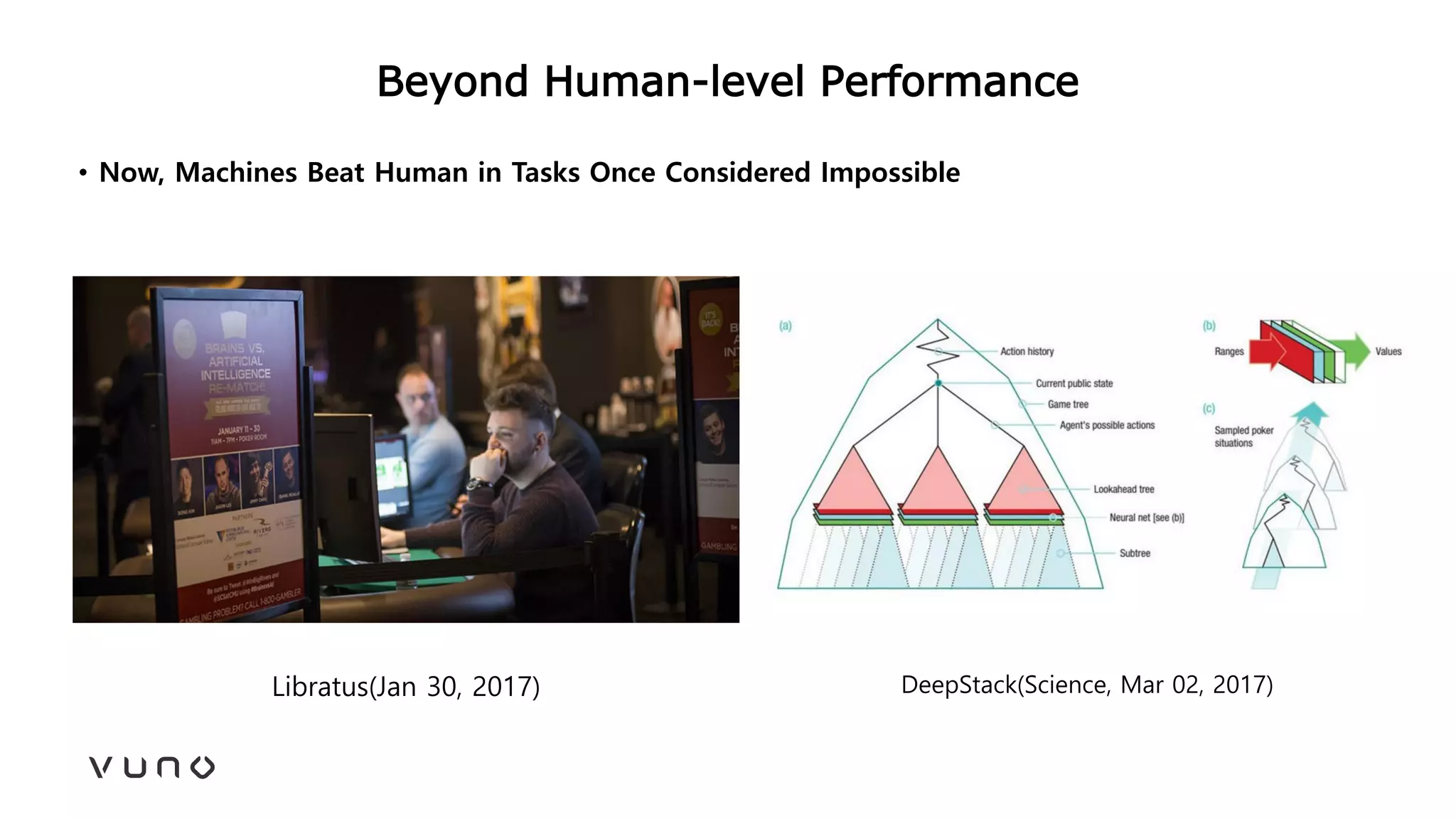
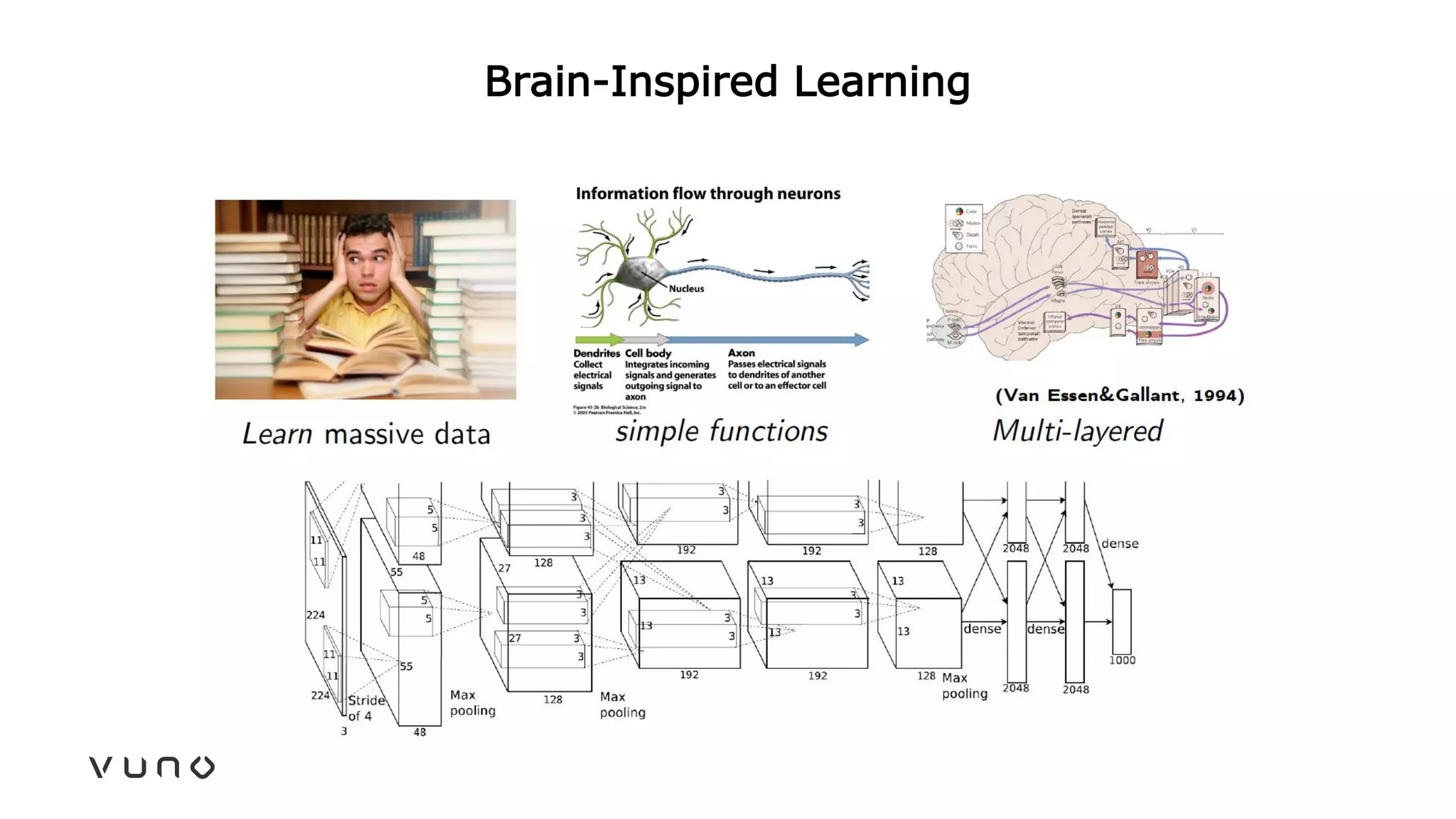
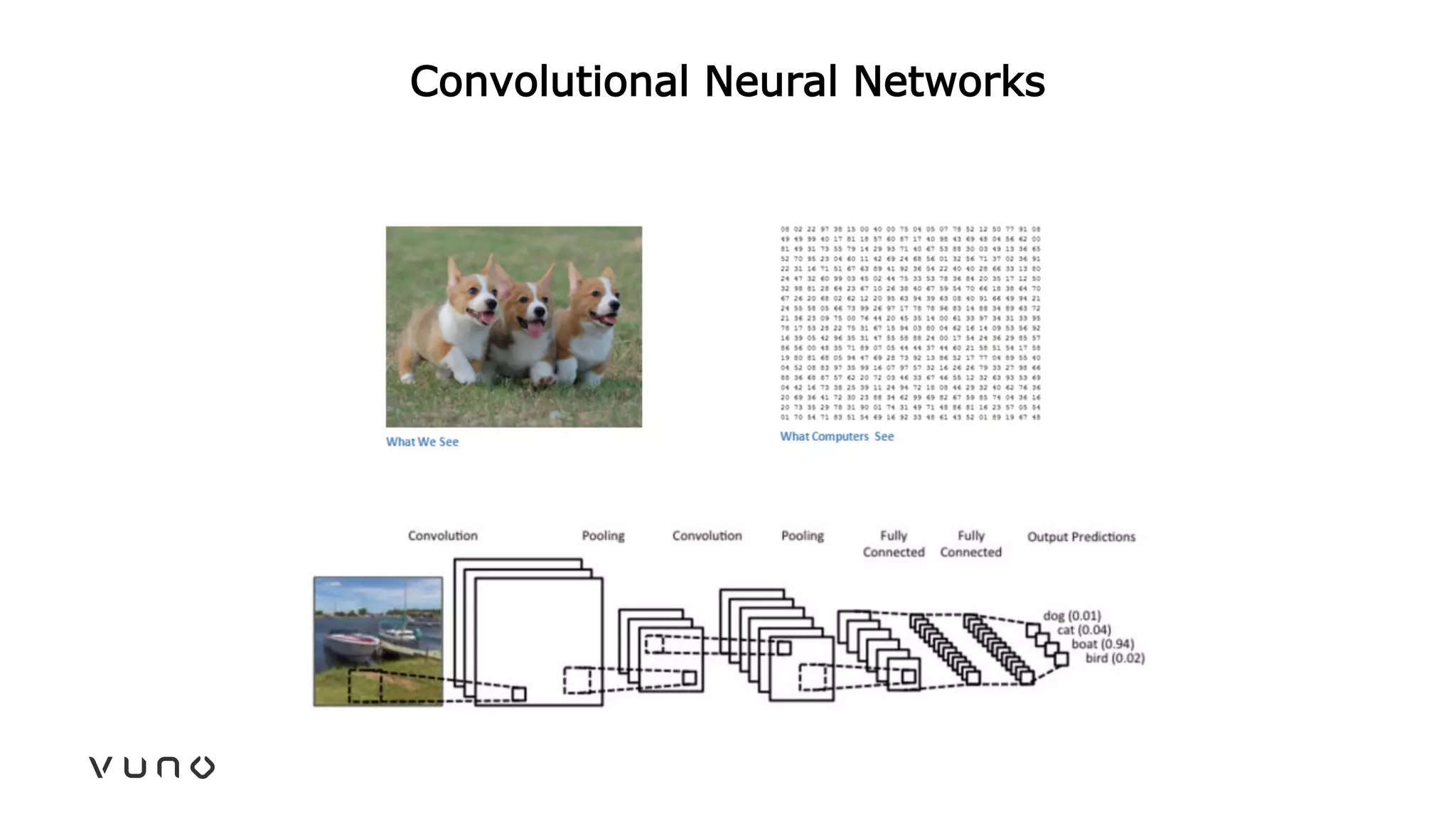
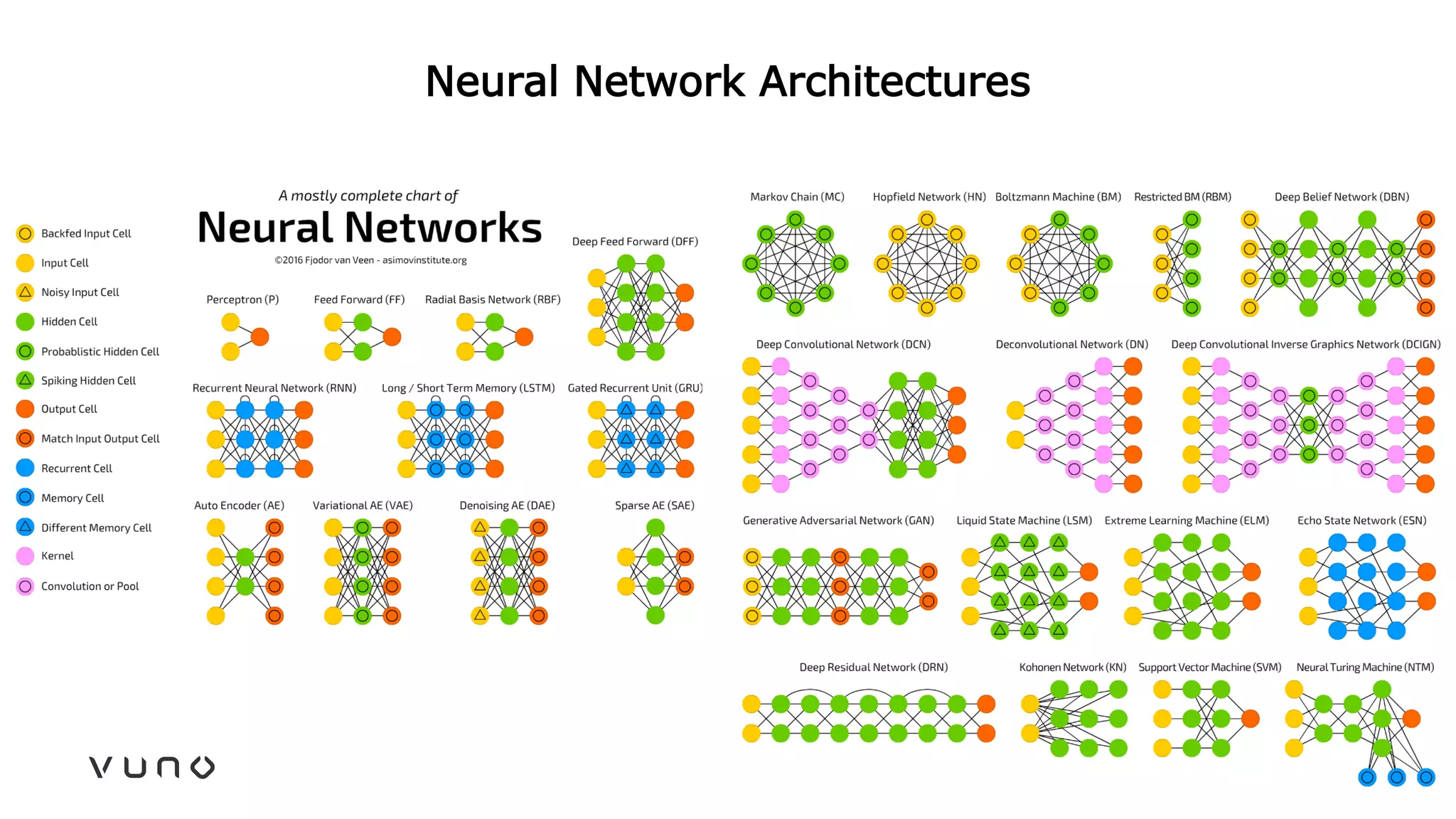
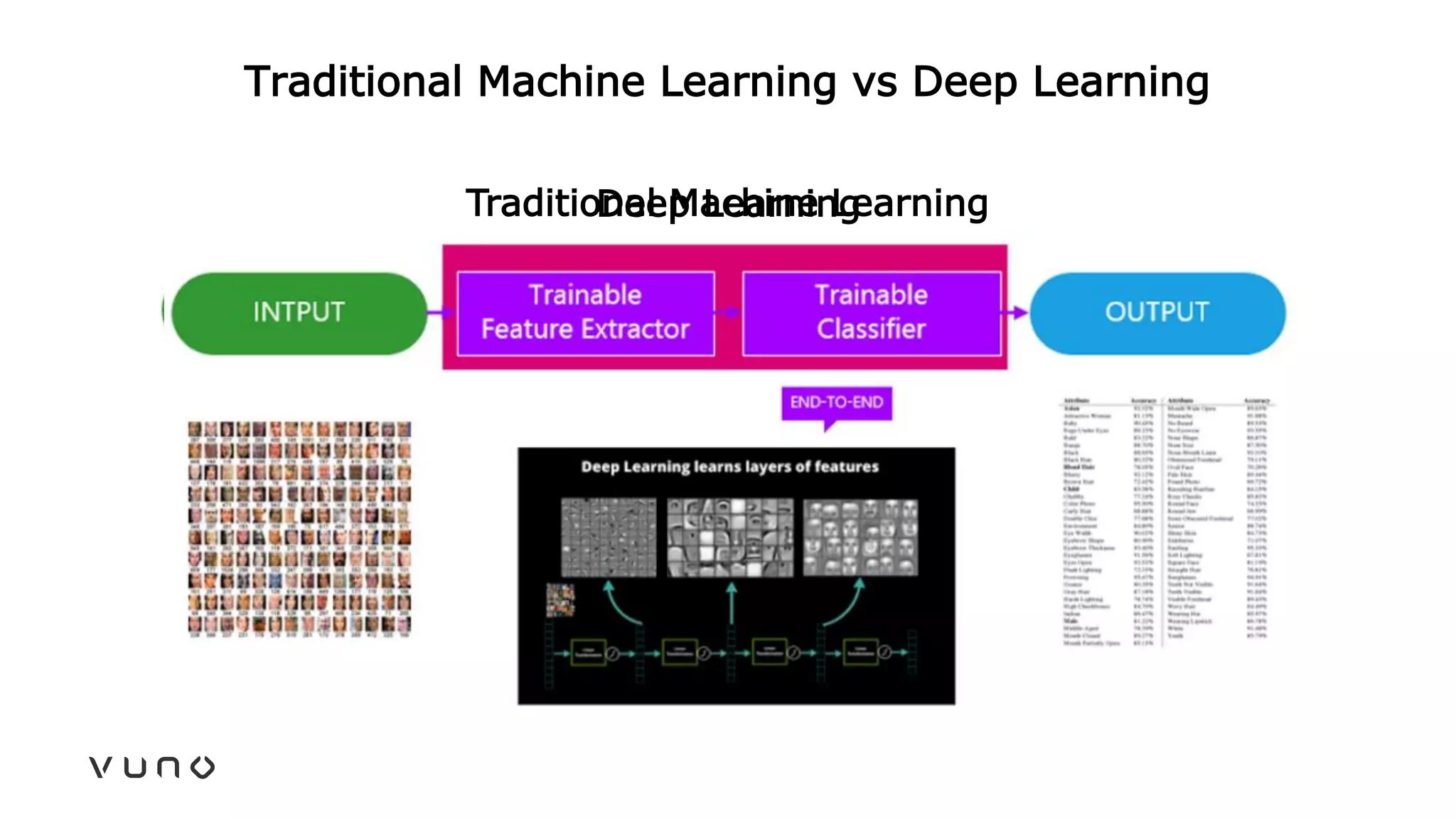
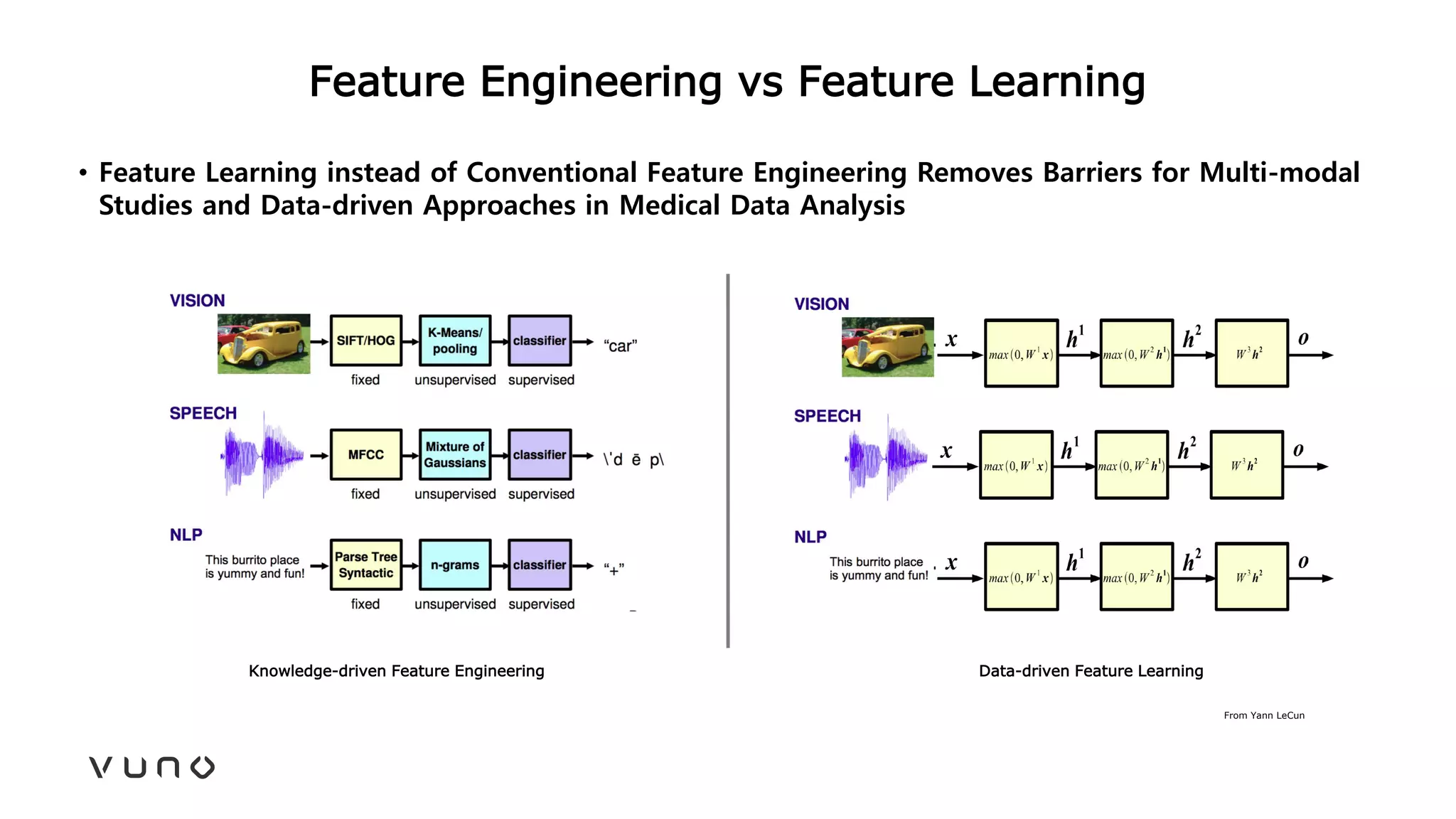
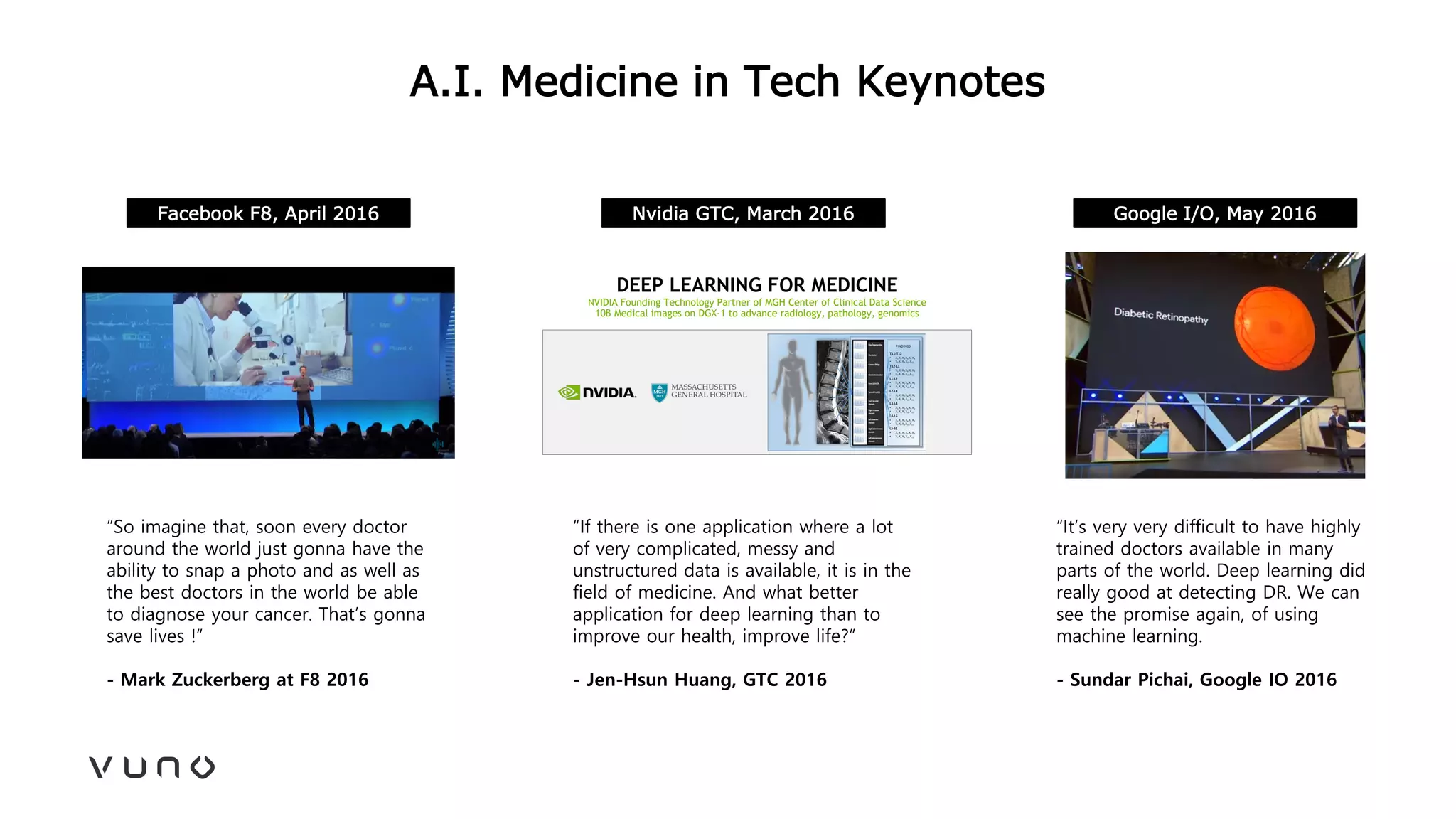
This document provides an overview of deep learning and its applications in medical imaging. It discusses key topics such as the definition of artificial intelligence, a brief history of neural networks and machine learning, and how deep learning is driving breakthroughs in tasks like visual and speech recognition. The document also addresses challenges in medical data analysis using deep learning, such as how to handle limited data or annotations. It provides examples of techniques used to address these challenges, such as data augmentation, transfer learning, and weakly supervised learning.






![AlphaGo in Medicine?
[Streams App for AKI Patient Management] [Medical Research]](https://image.slidesharecdn.com/practicalpointsofdeeplearningformedicalimaging-170611182119/75/2017-06-Practical-points-of-deep-learning-for-medical-imaging-28-2048.jpg)