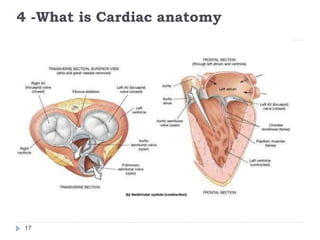
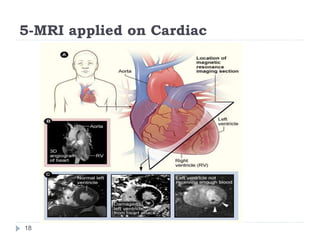
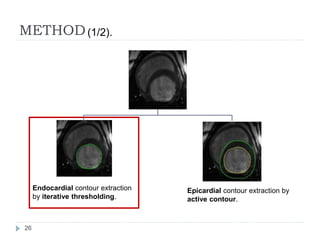
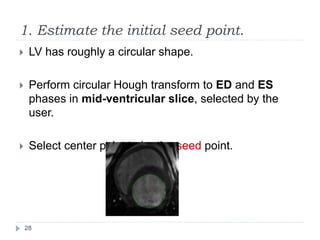
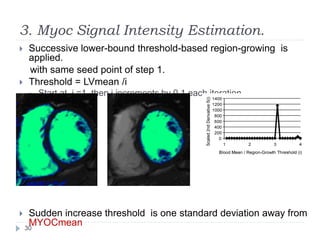
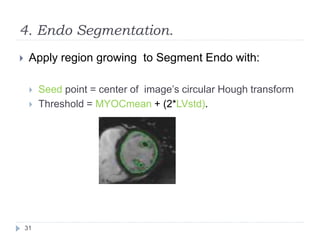
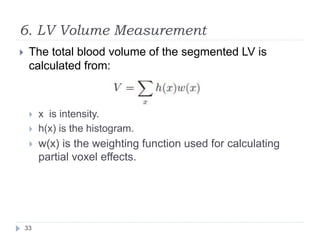
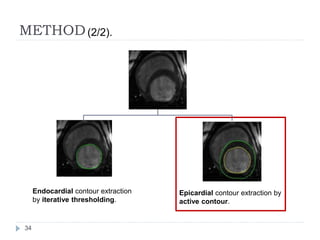
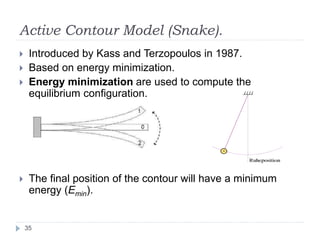
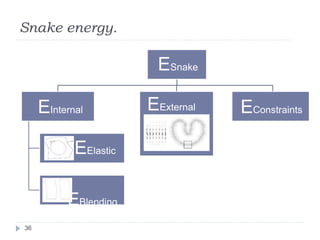
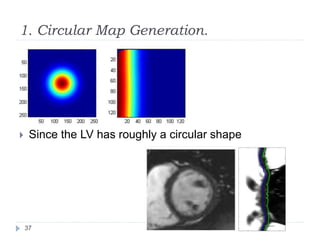
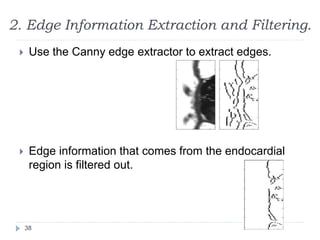
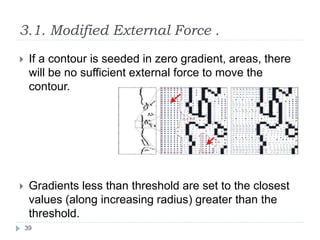
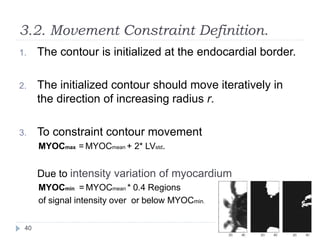
This document summarizes an automatic left ventricle segmentation technique using iterative thresholding and an active contour model adapted for short-axis cardiac MRI images. It begins with background on image segmentation and its applications. Then, it reviews related work on cardiac segmentation techniques and their limitations. The proposed method segments the endocardium using iterative thresholding and the epicardium using an active contour model. It estimates blood and myocardial intensities, applies region growing to segment the endocardium in each slice, and propagates the segmentation to remaining slices. Finally, it measures left ventricle volume and compares the results to manual segmentation.