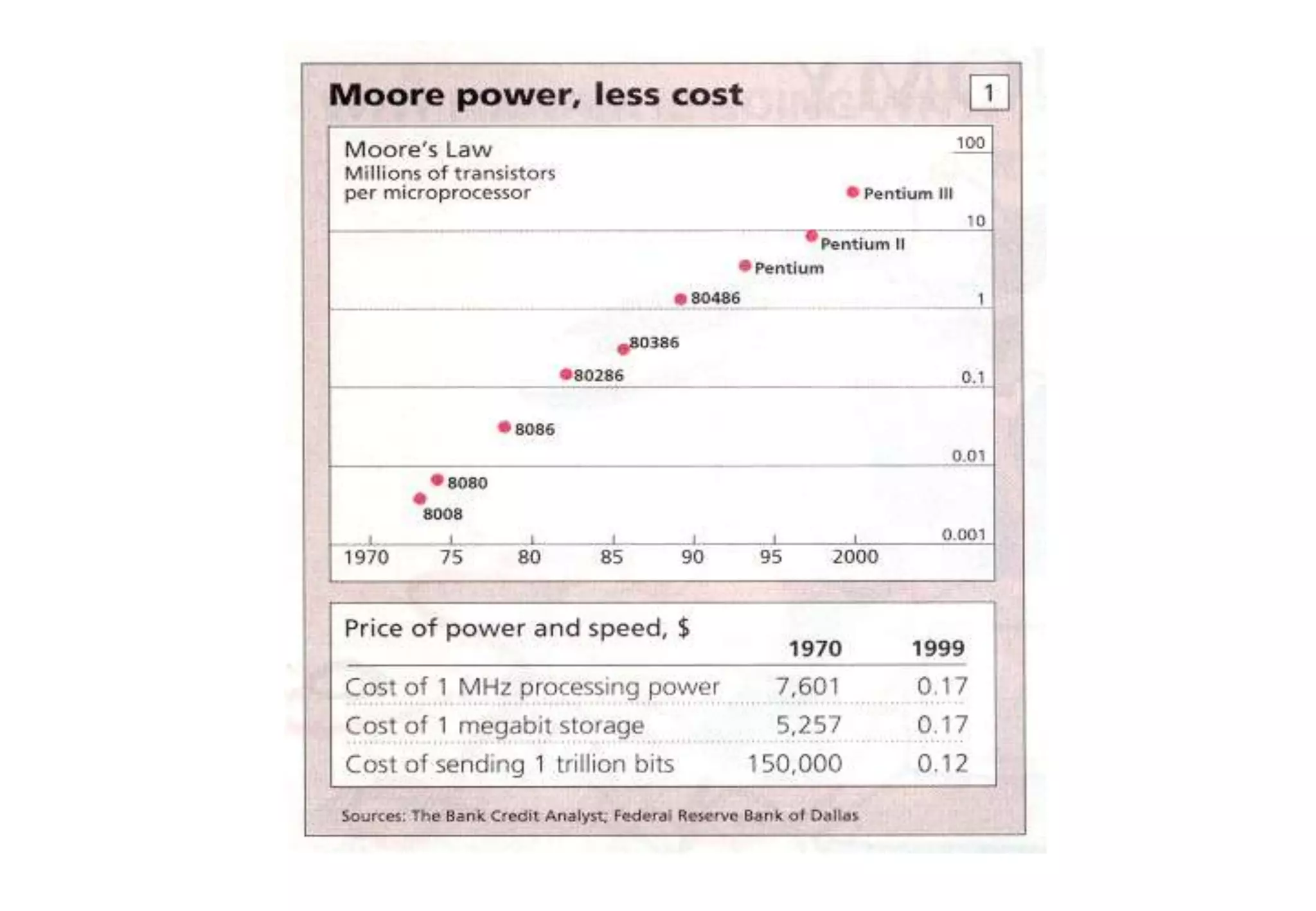
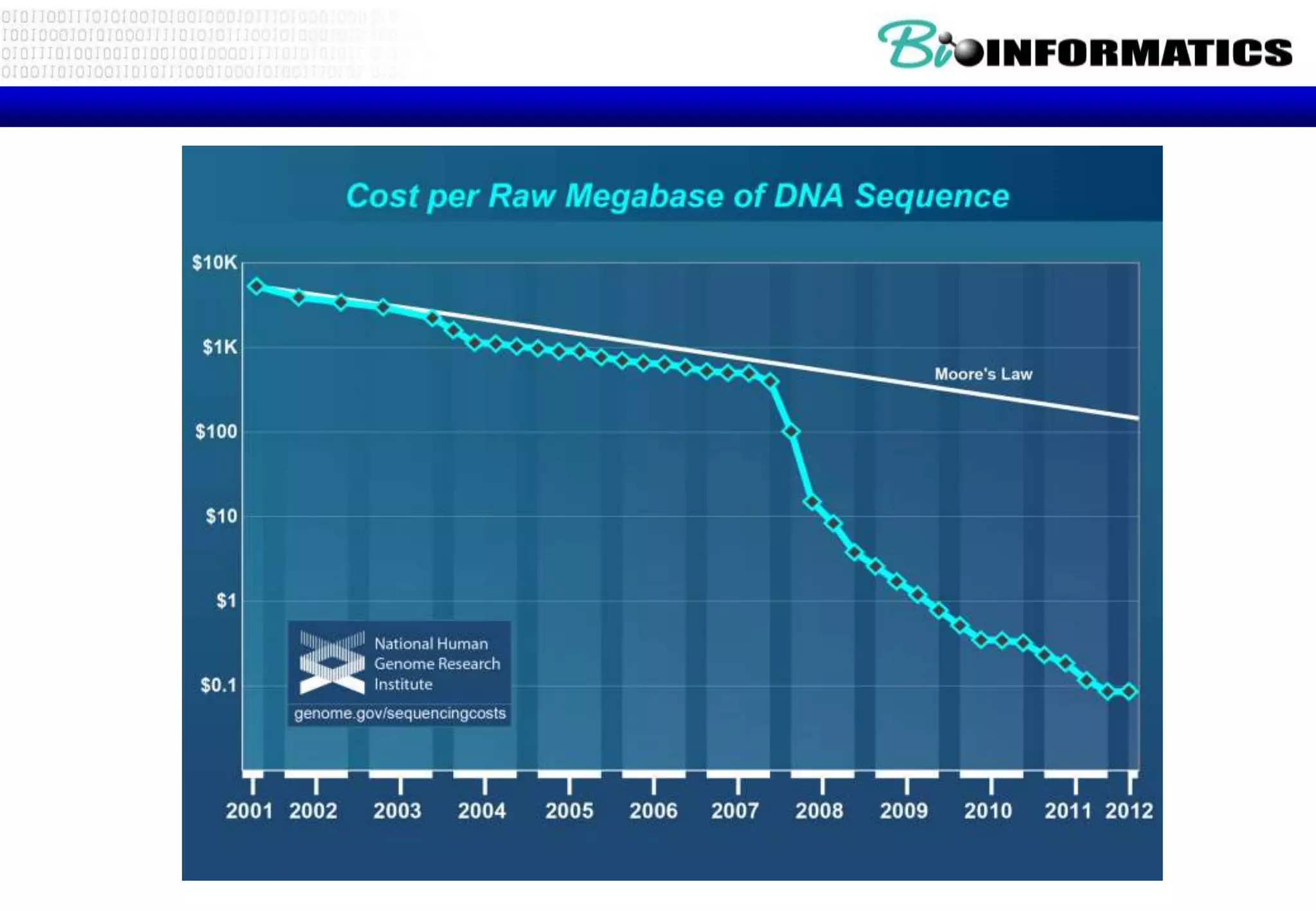
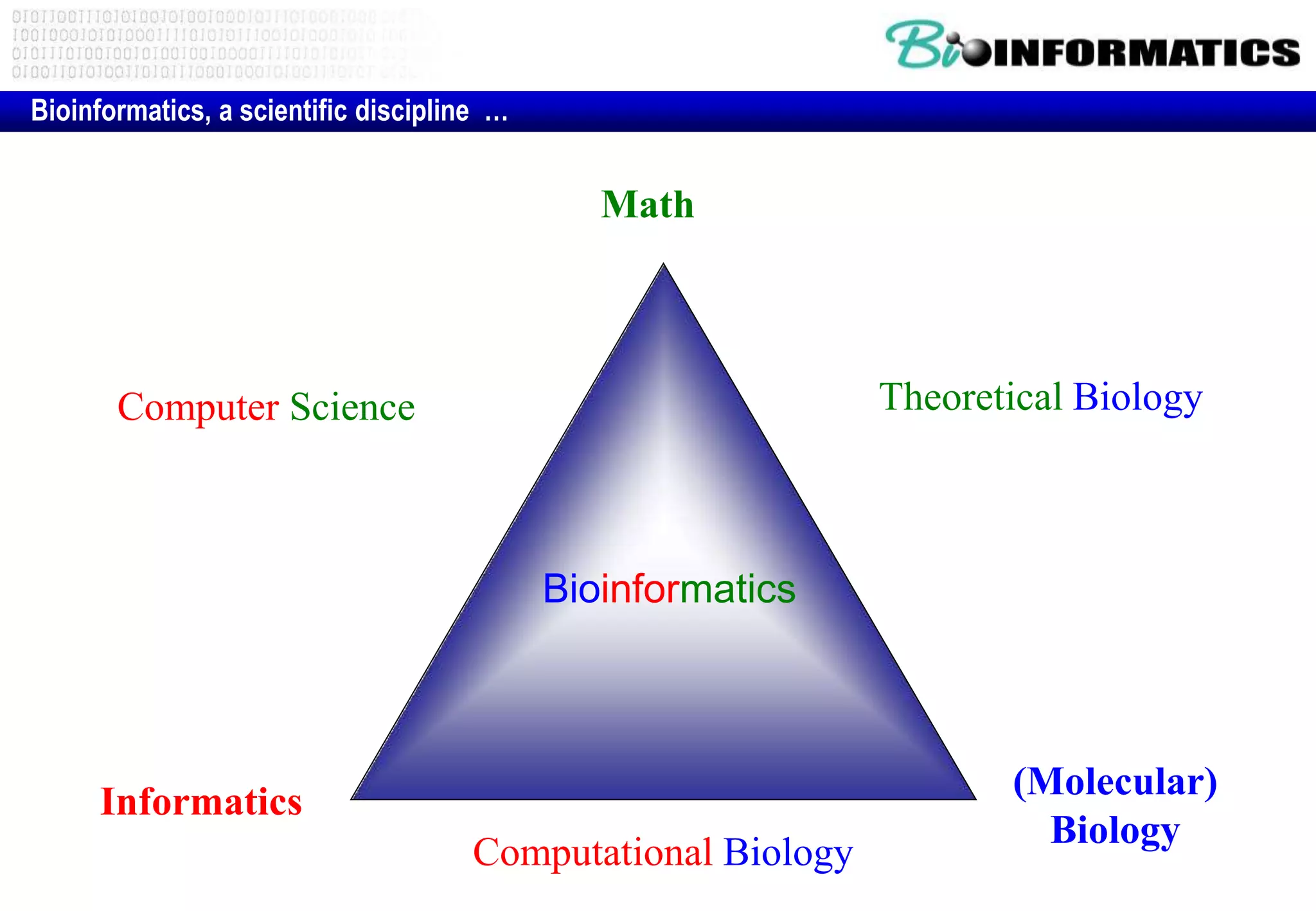
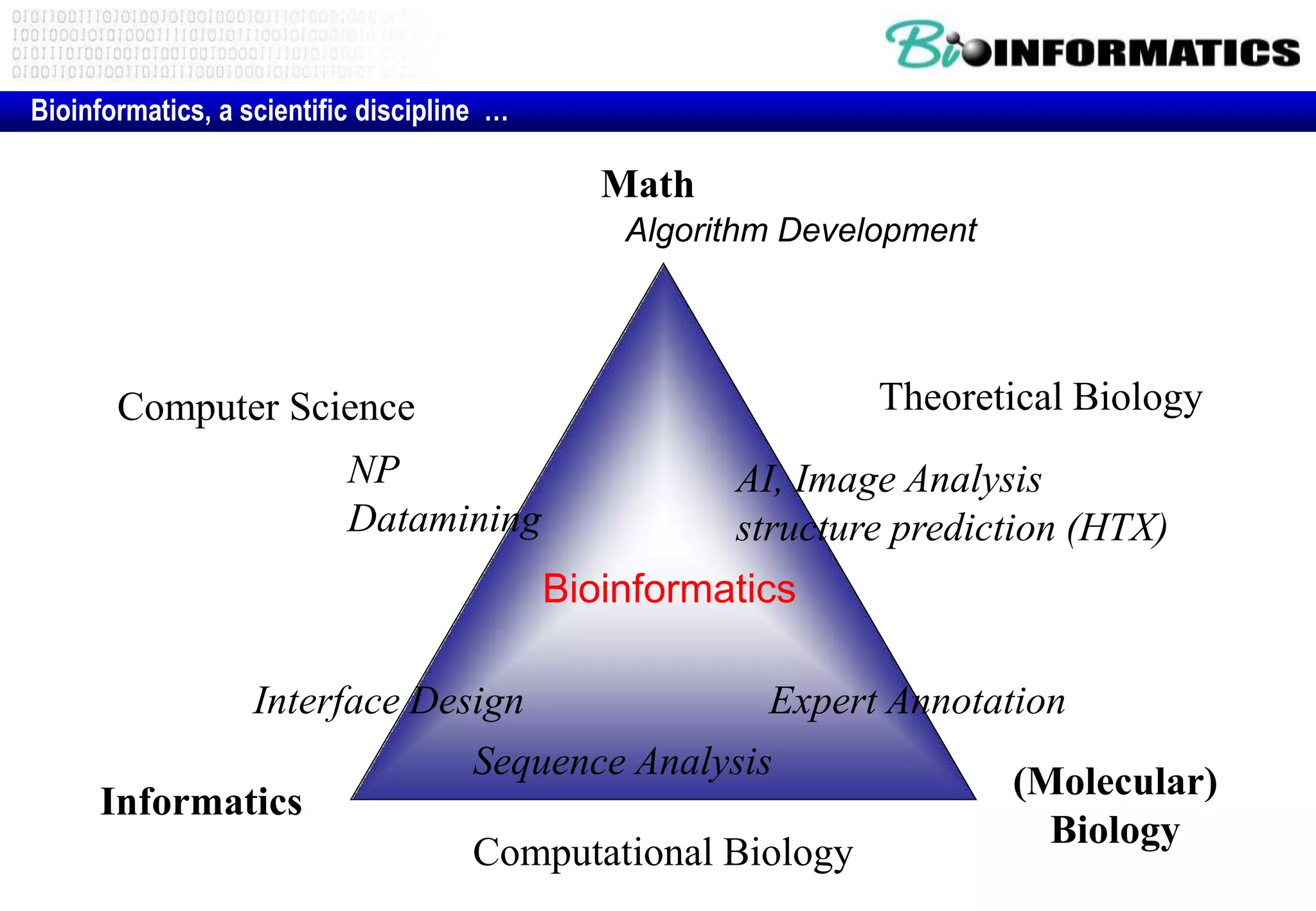
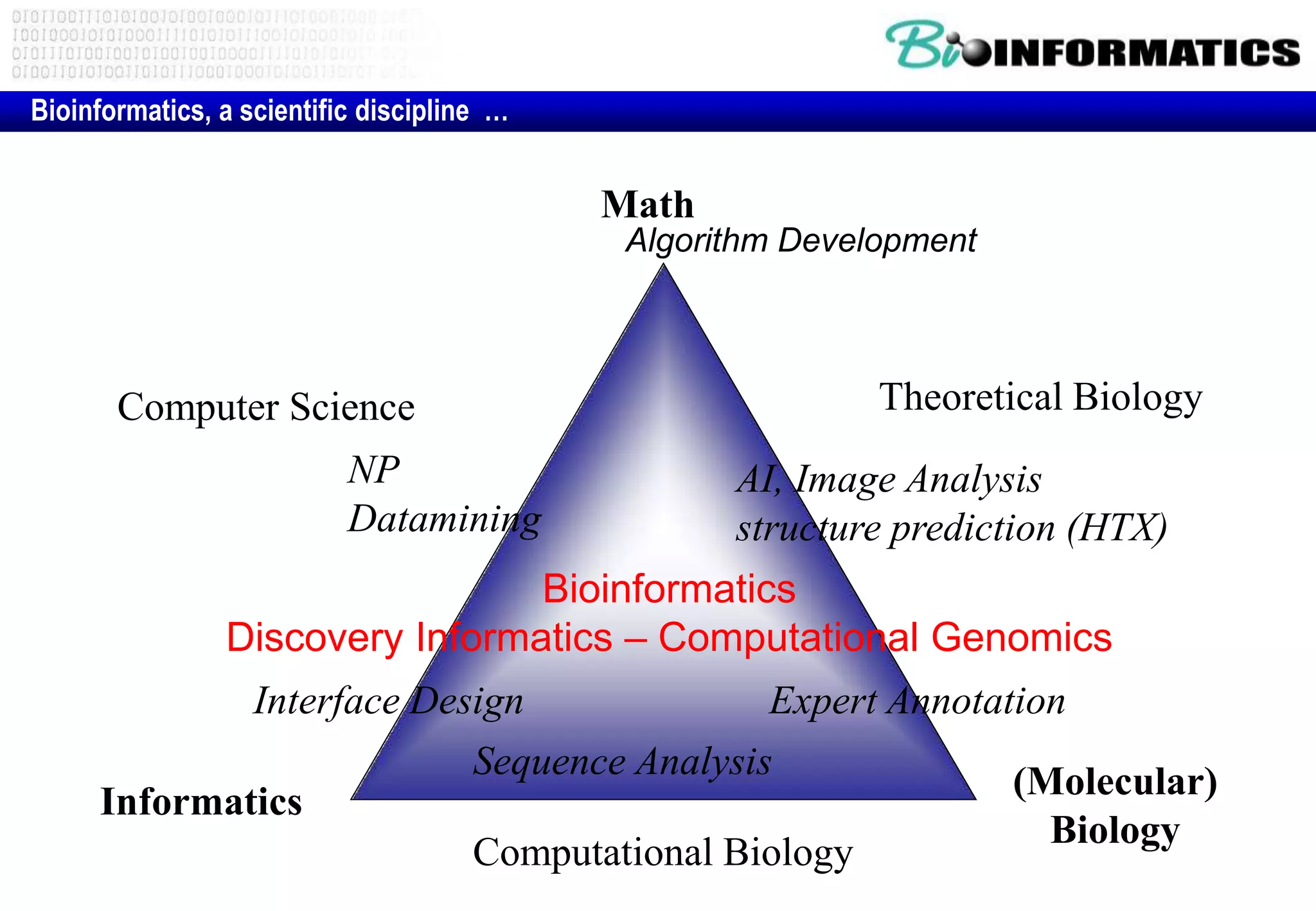
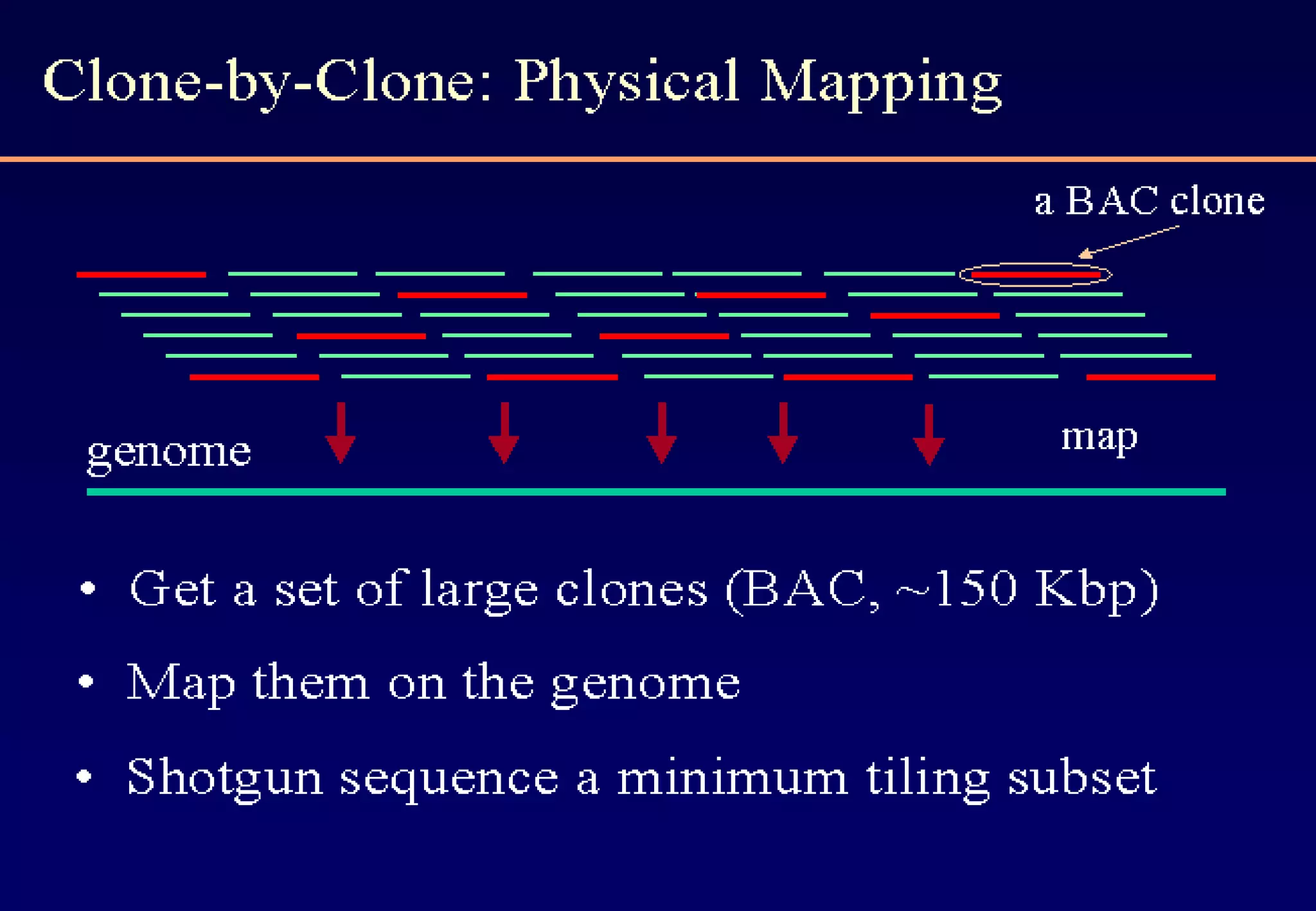
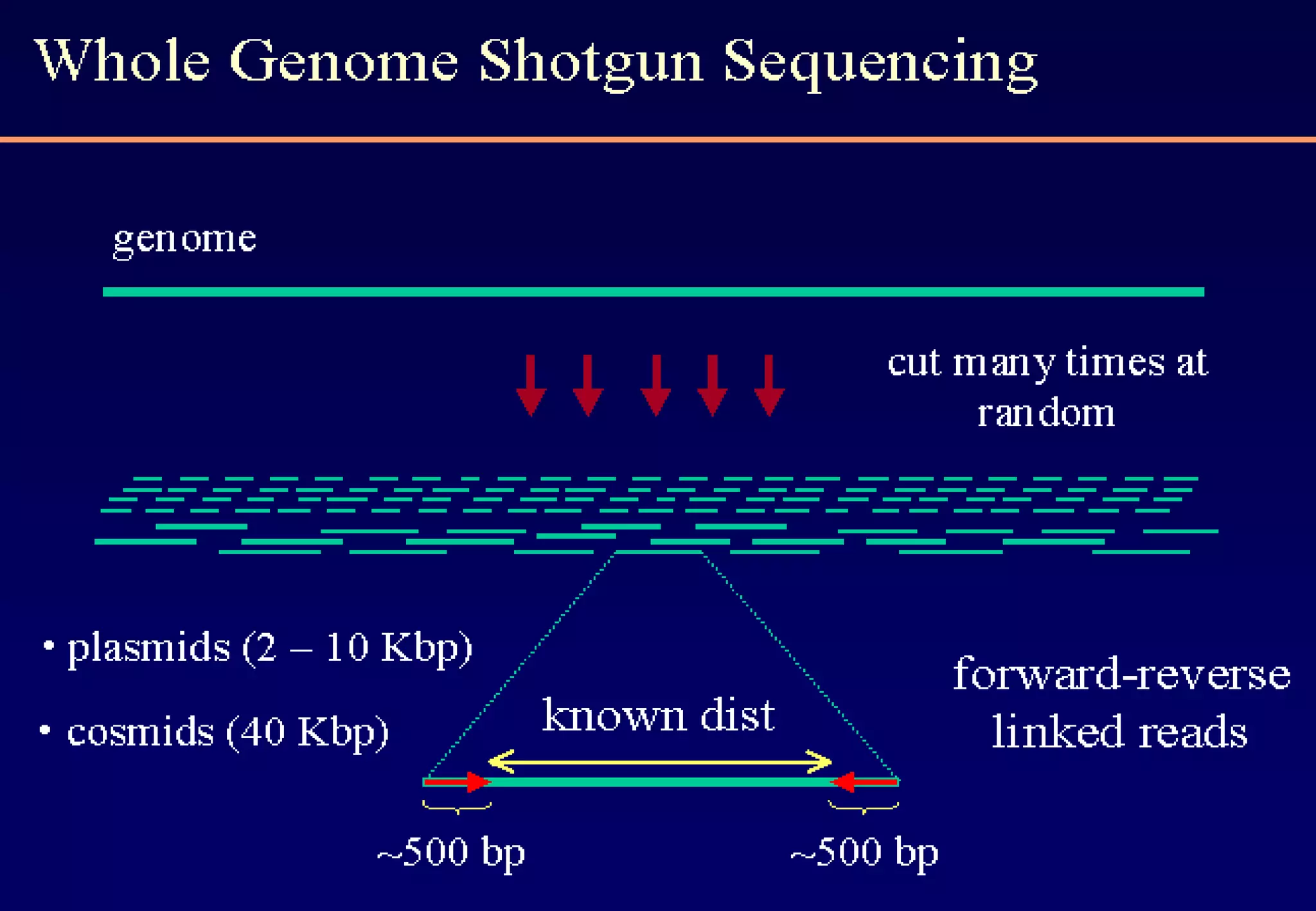
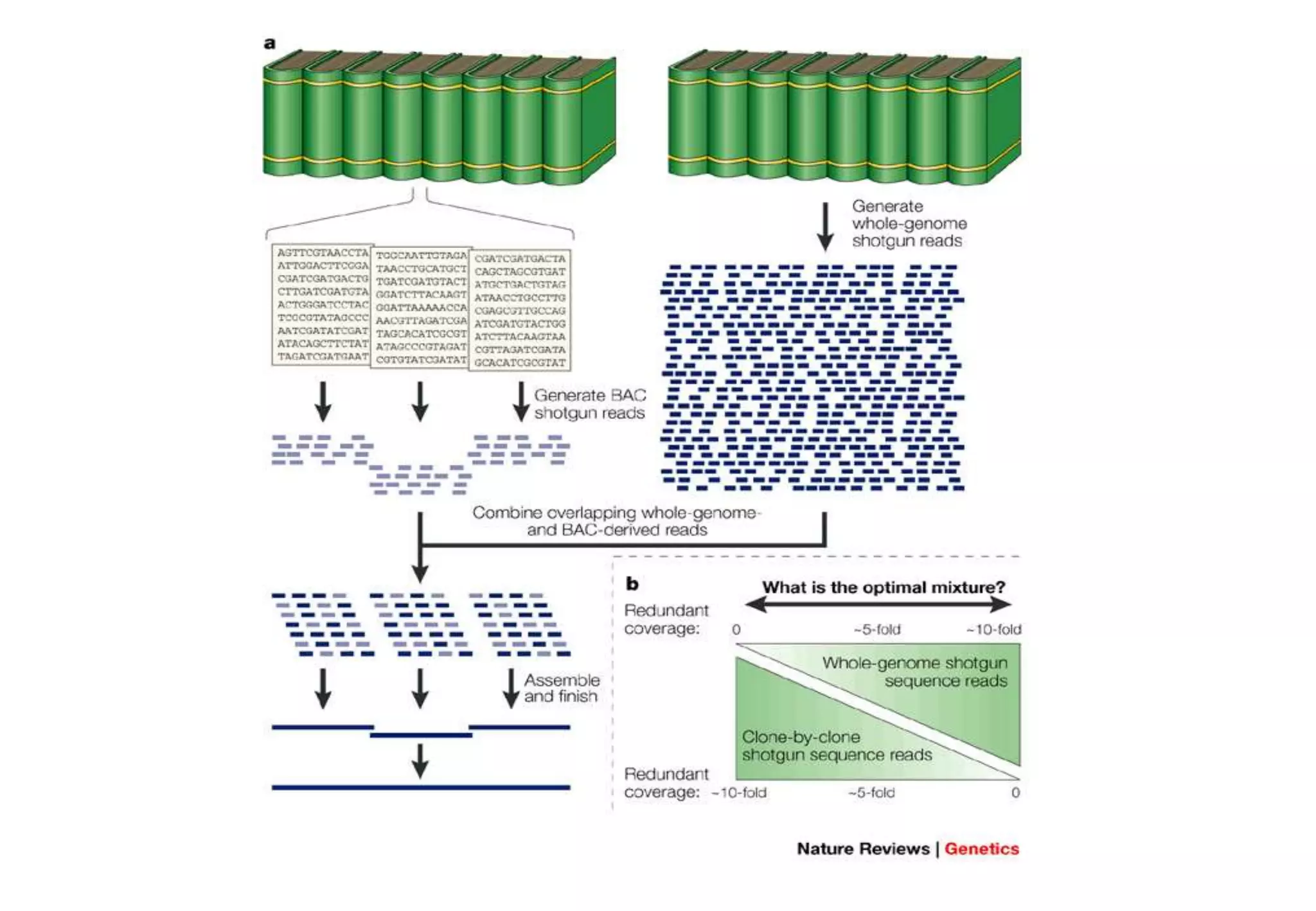
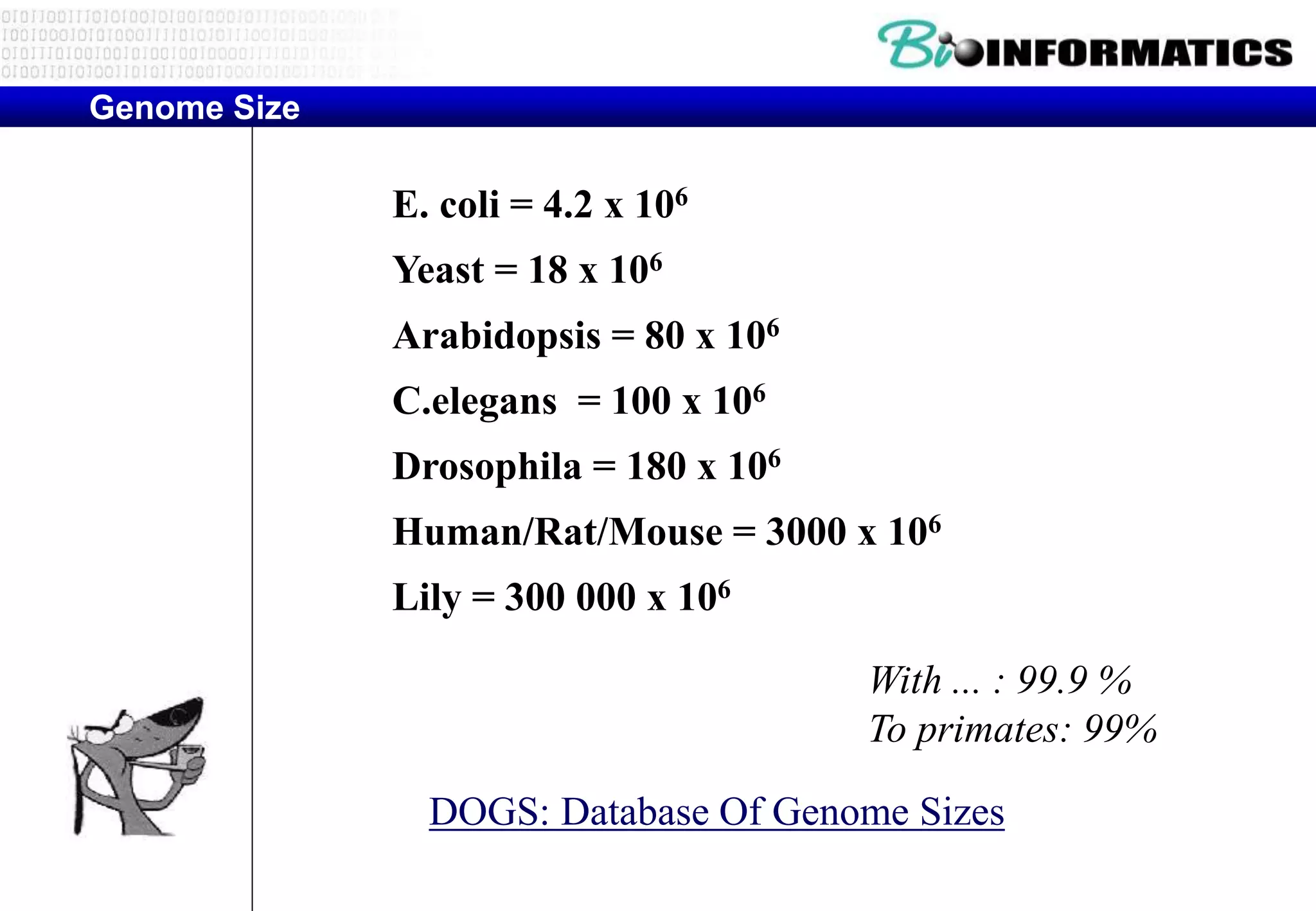
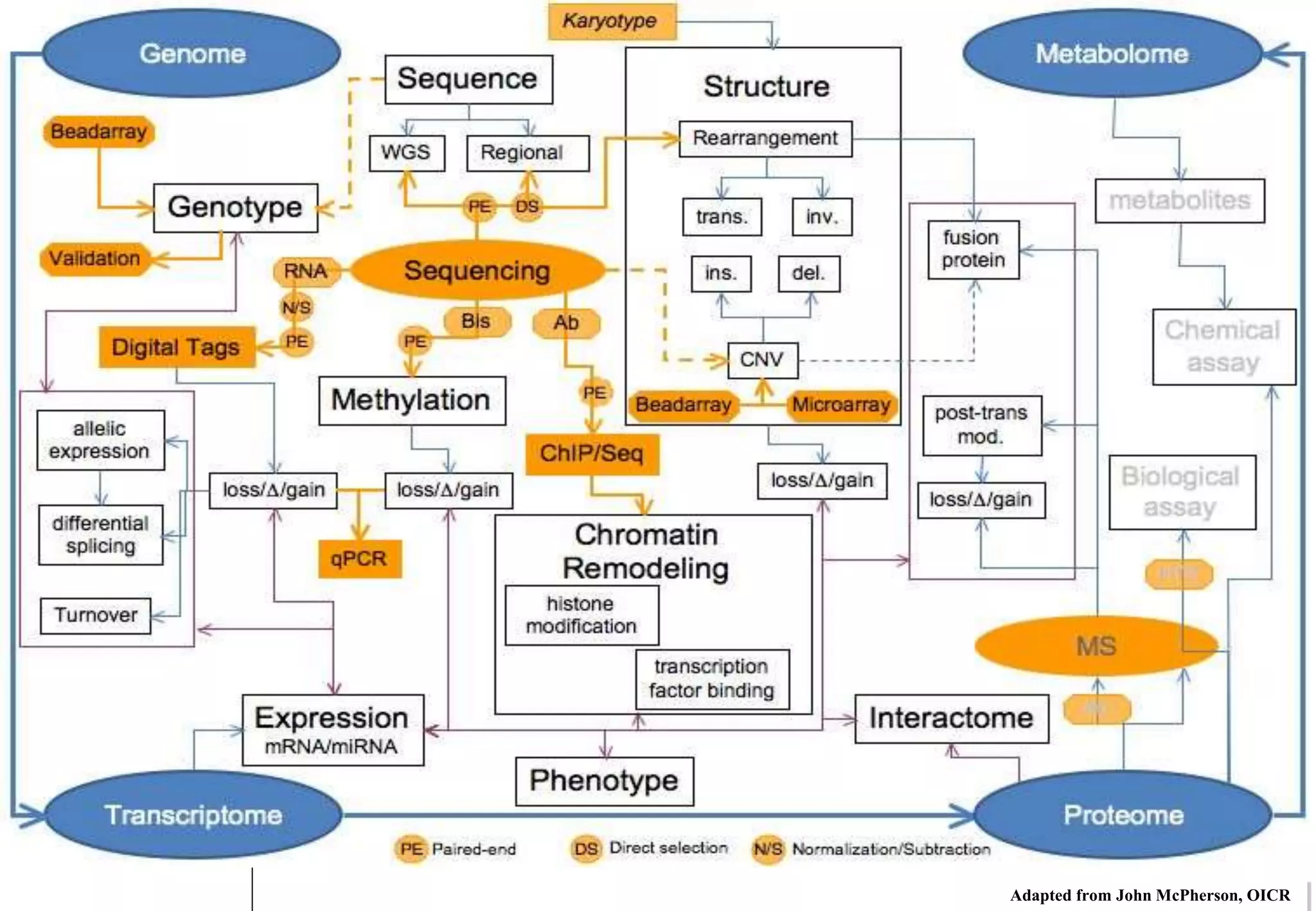
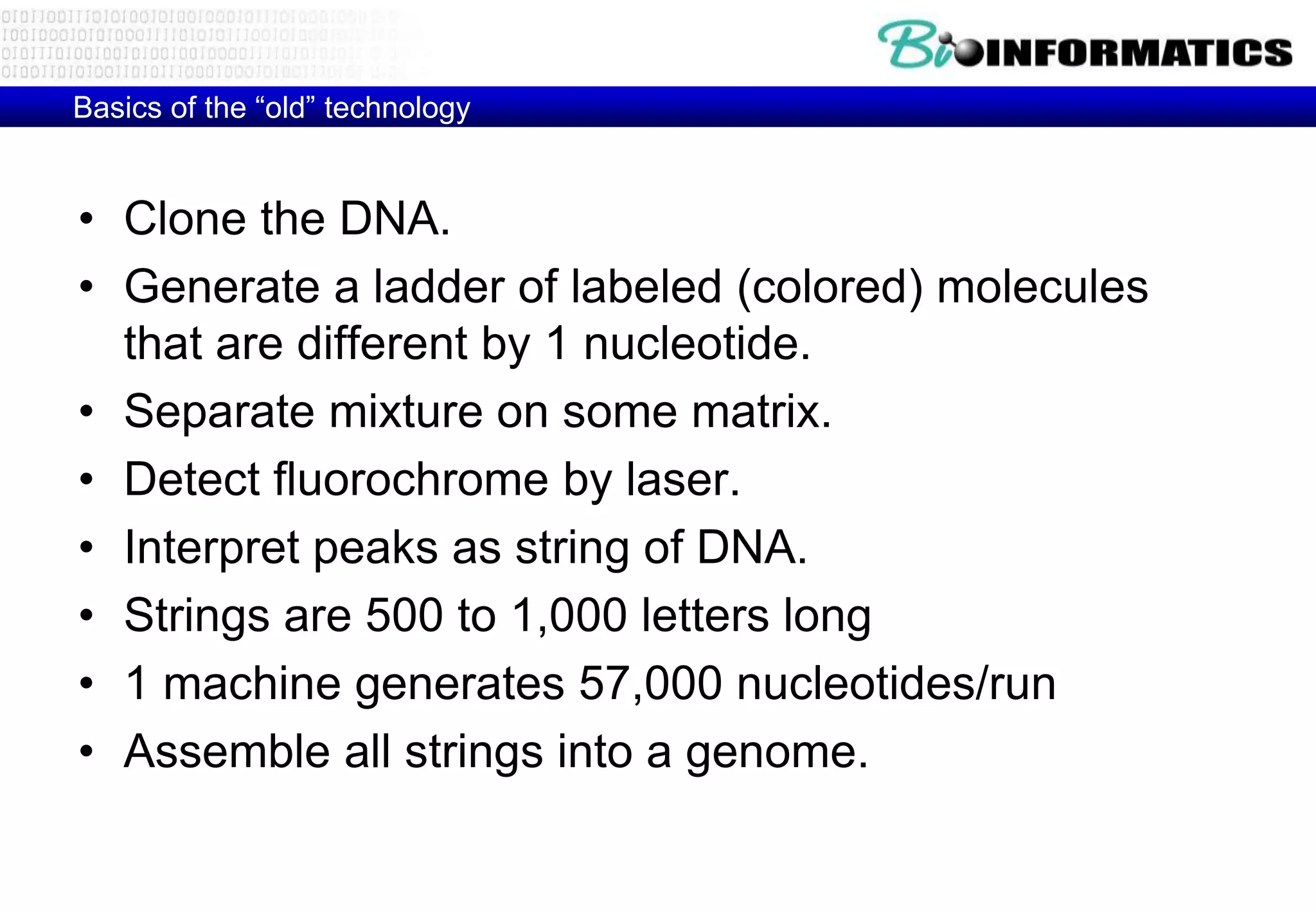
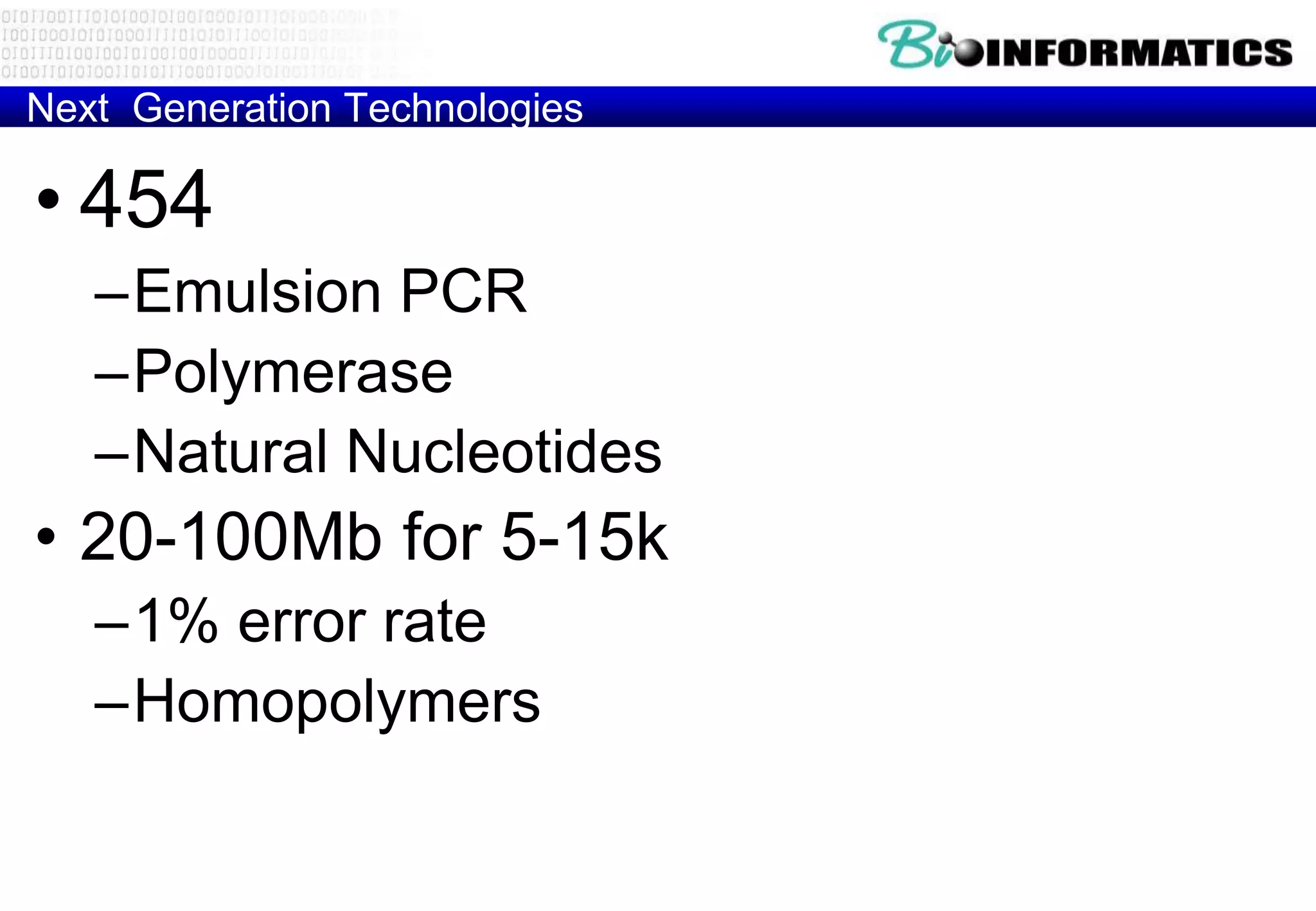
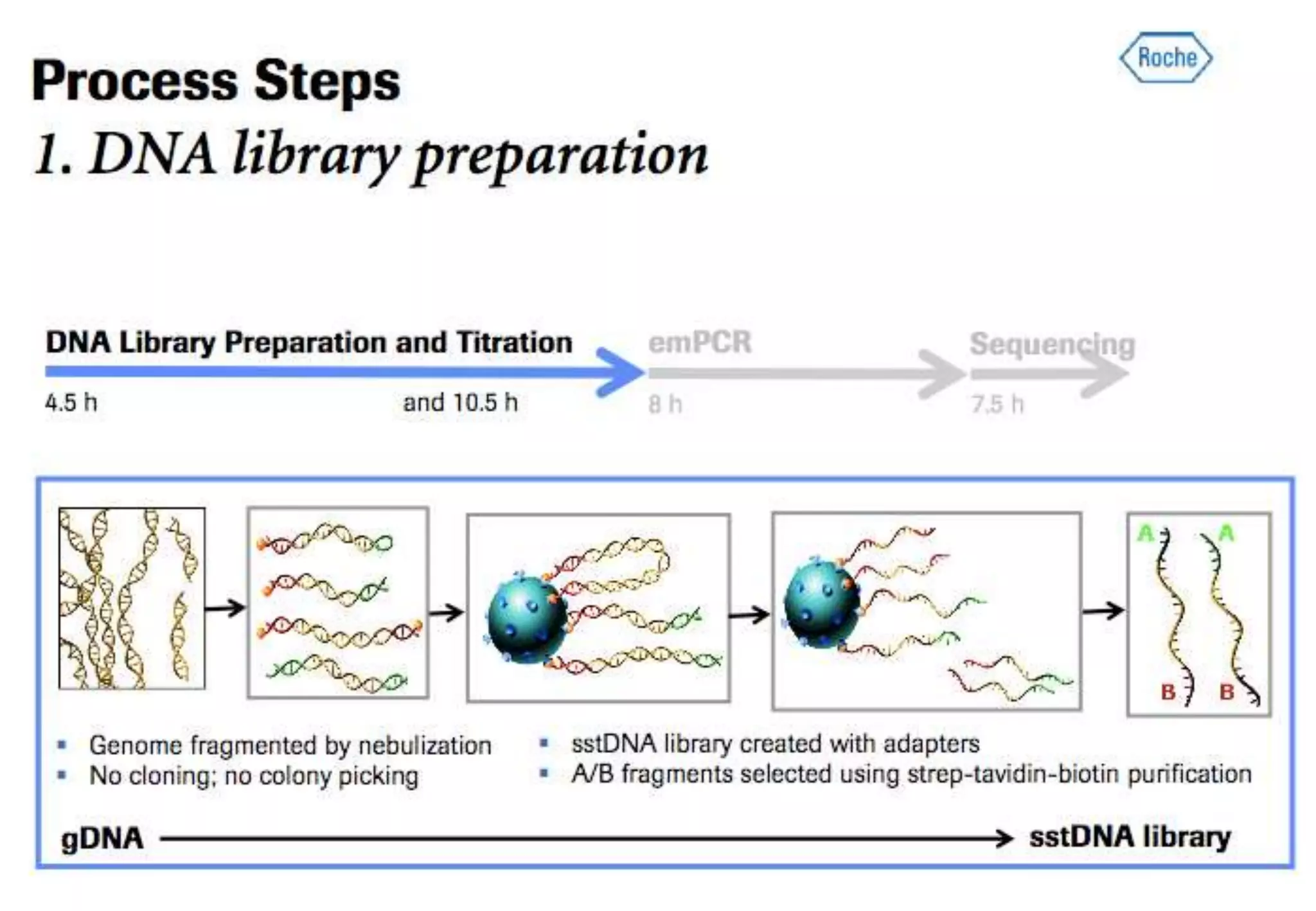
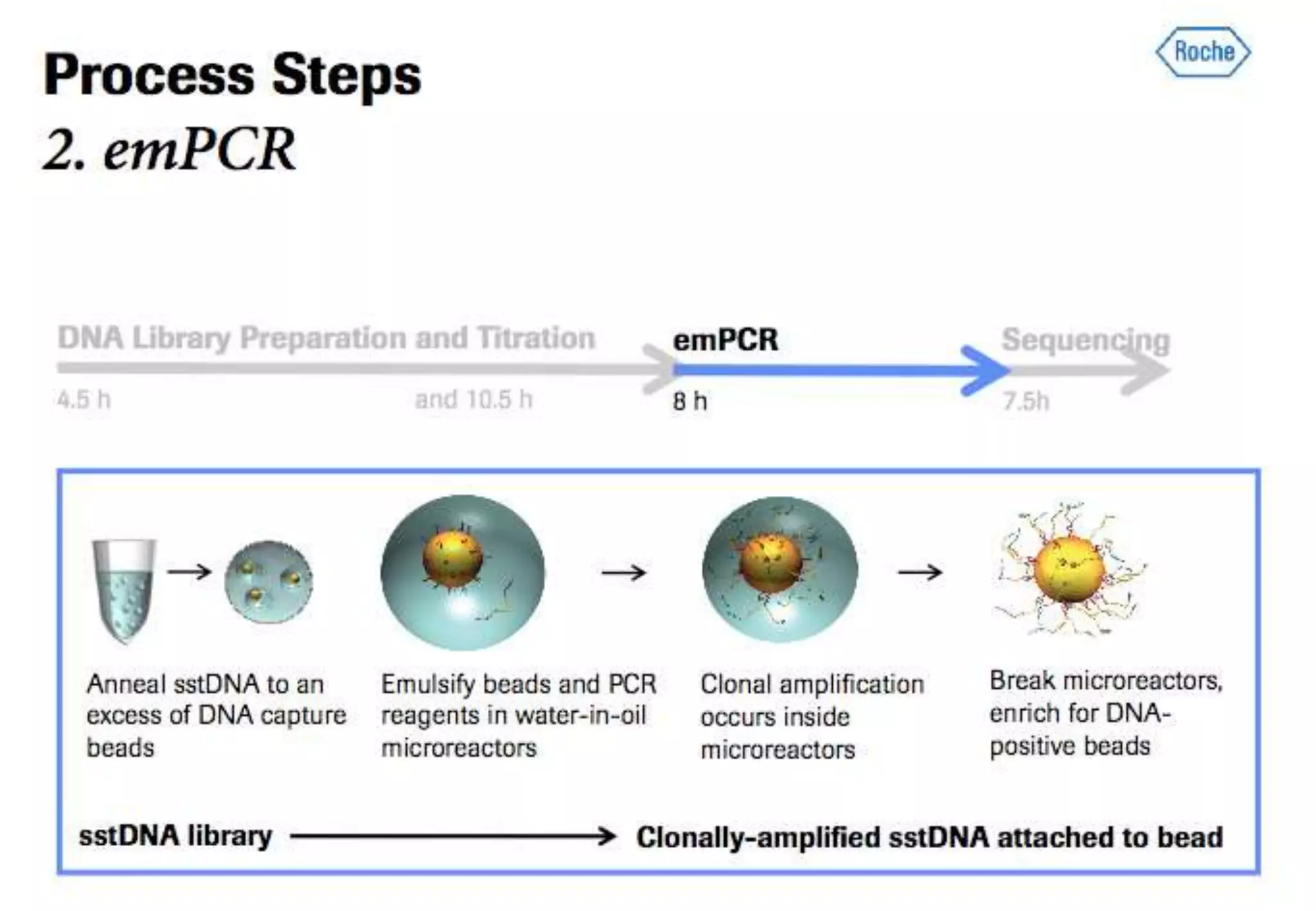
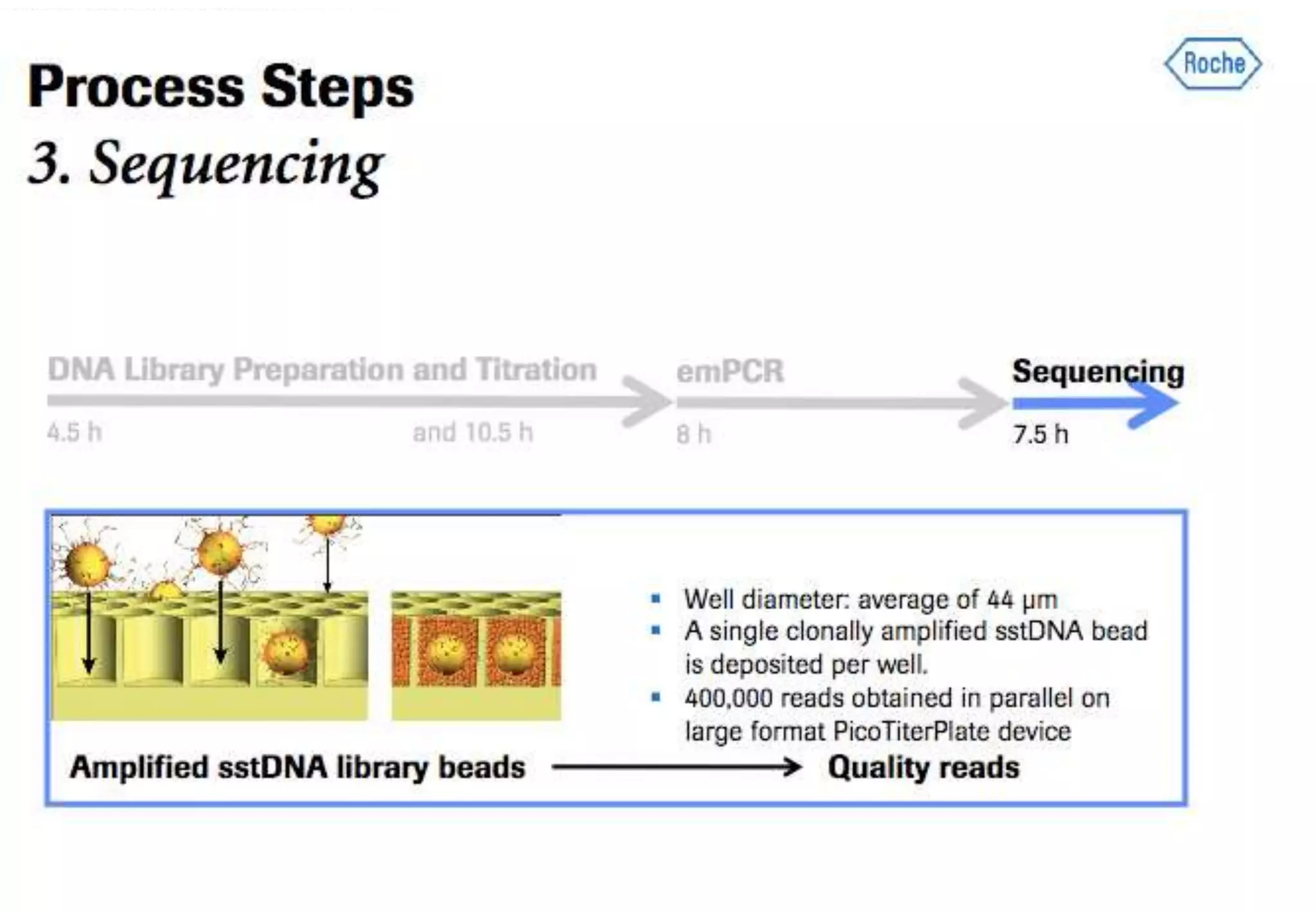
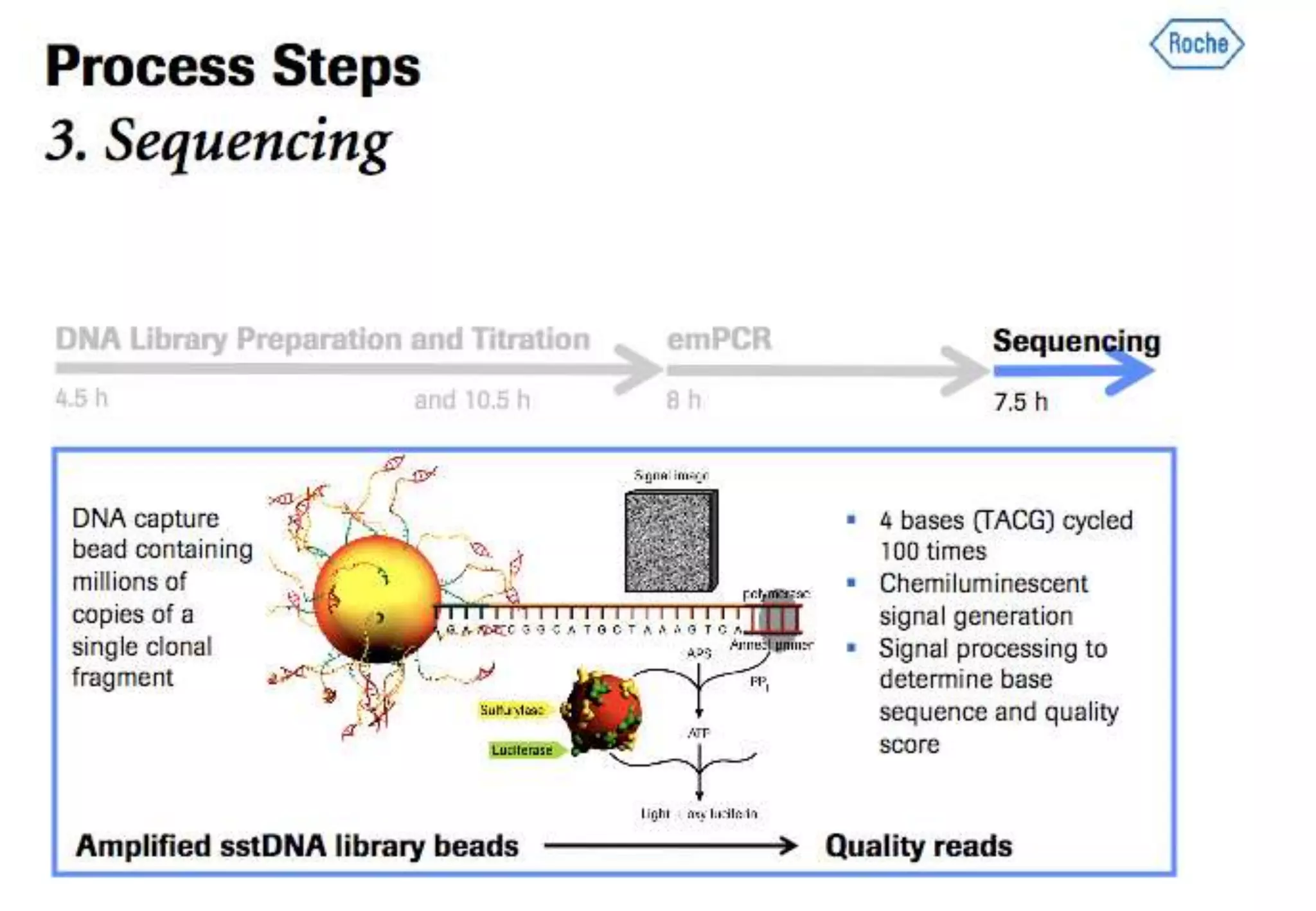
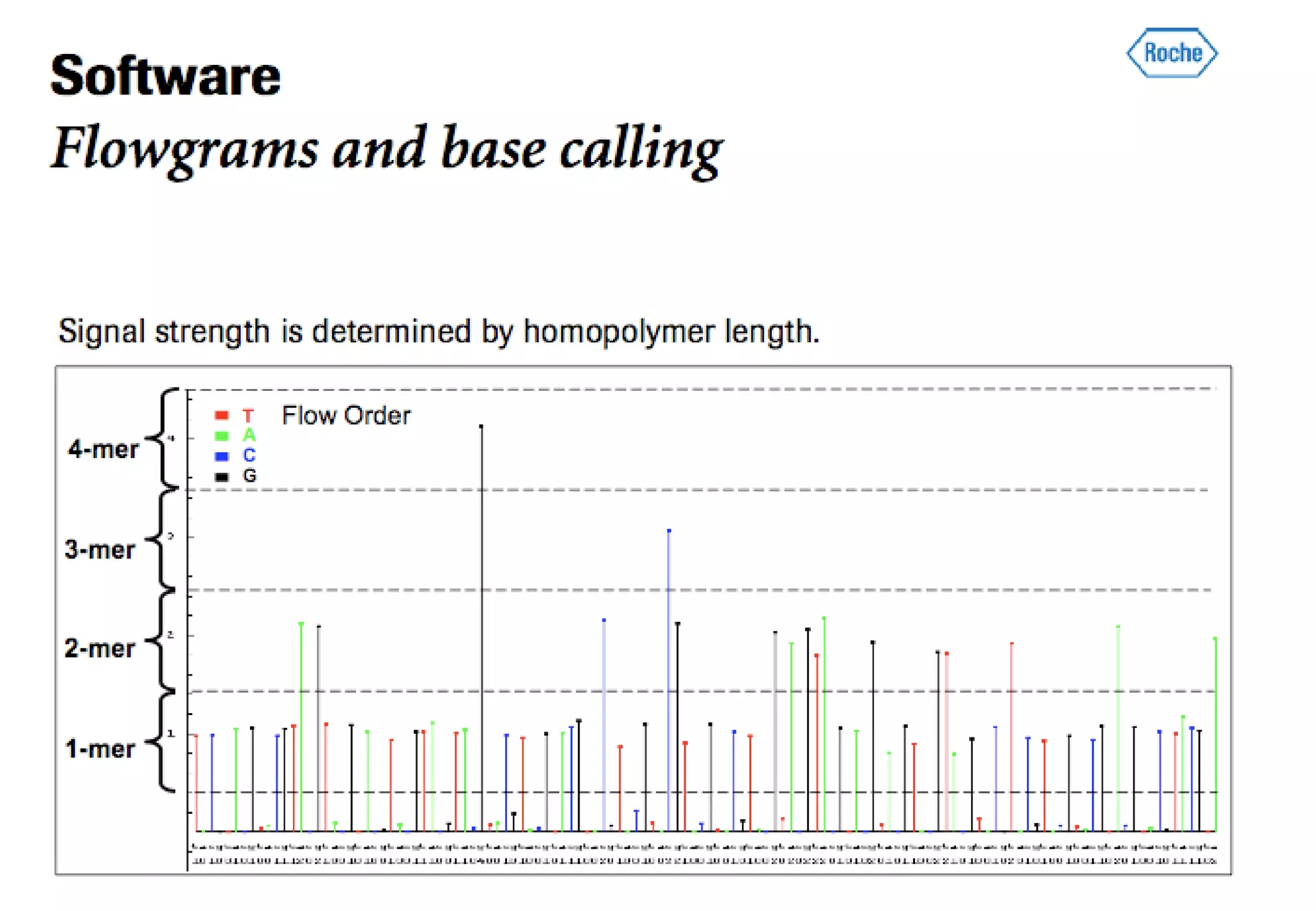
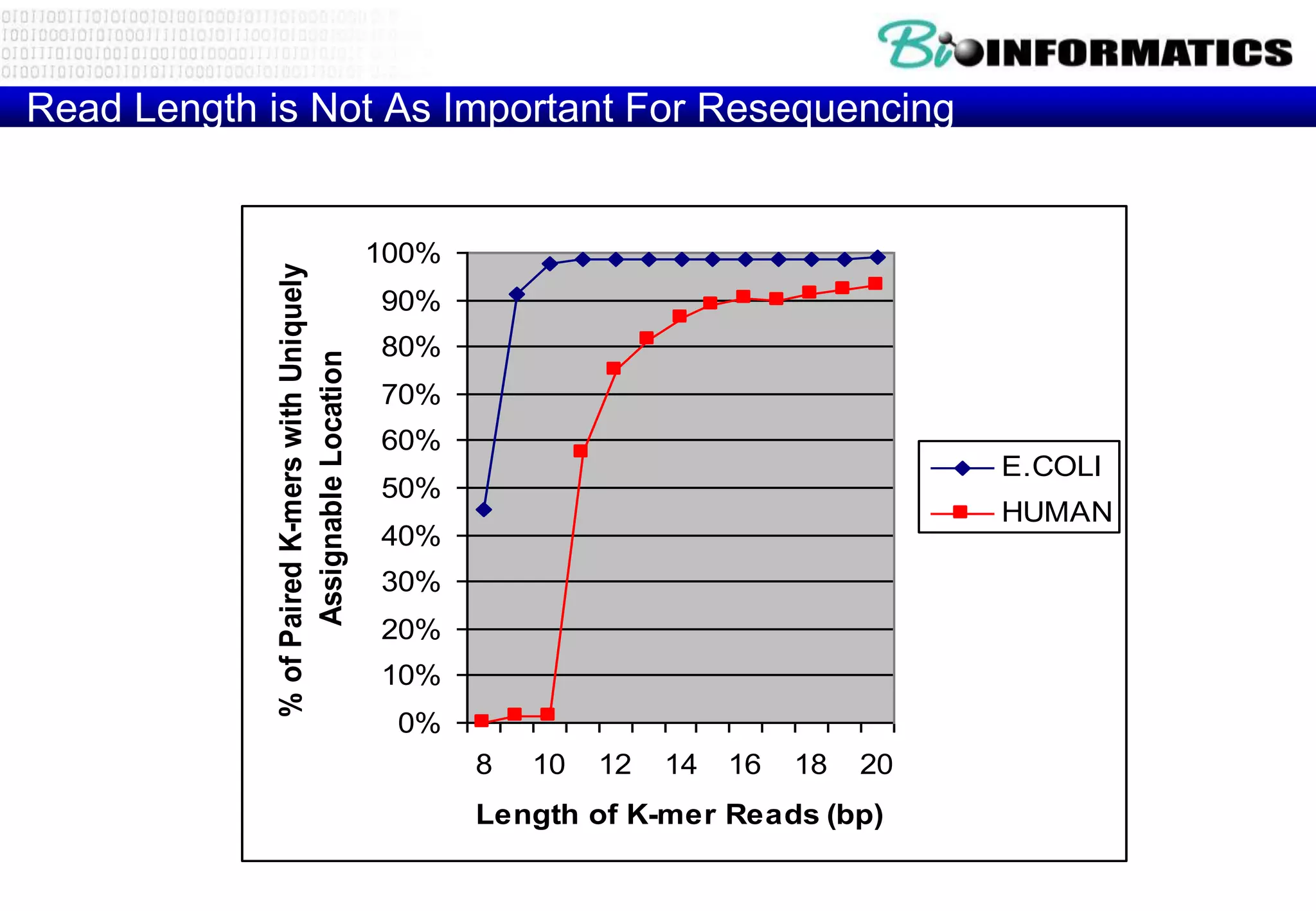
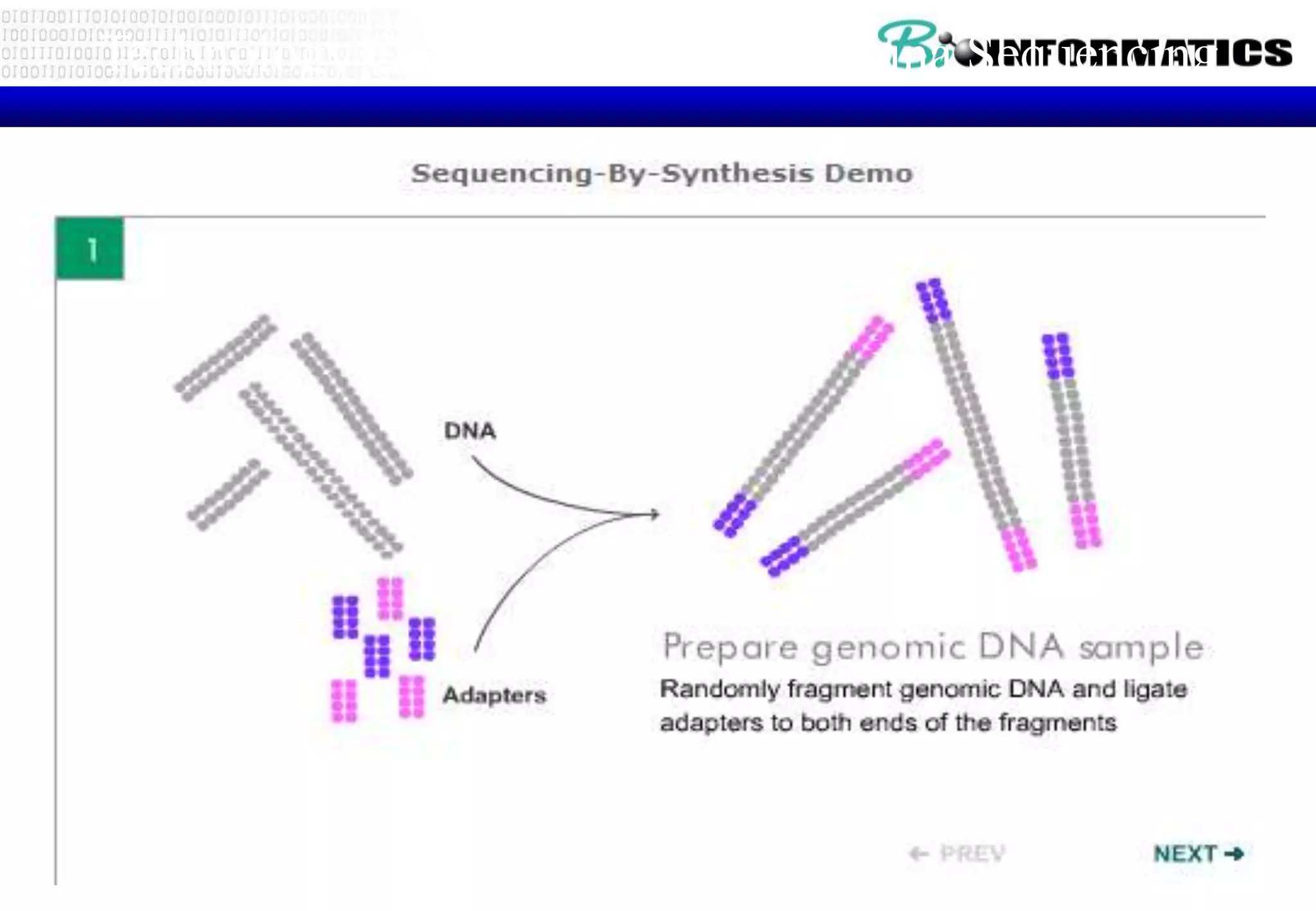
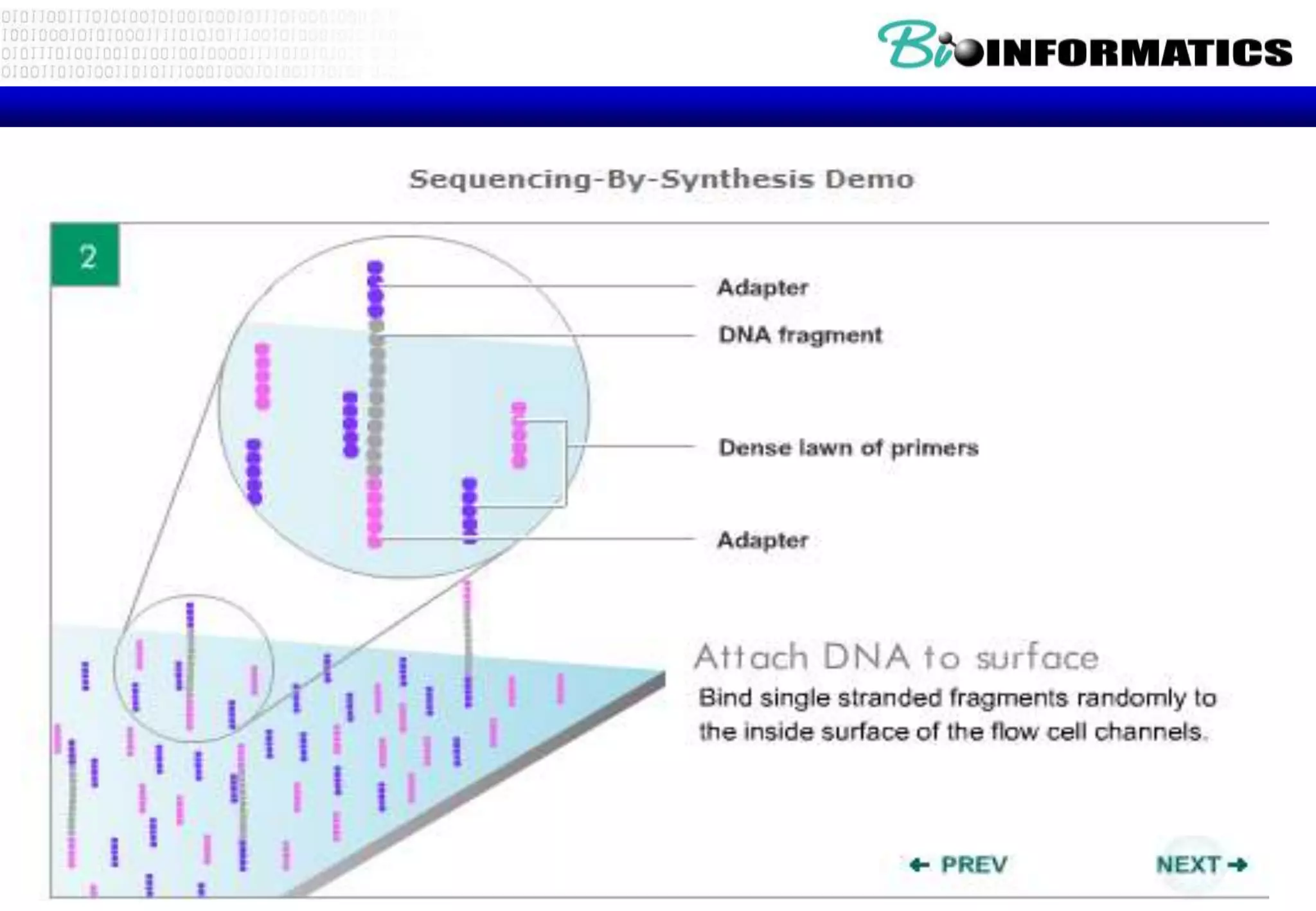
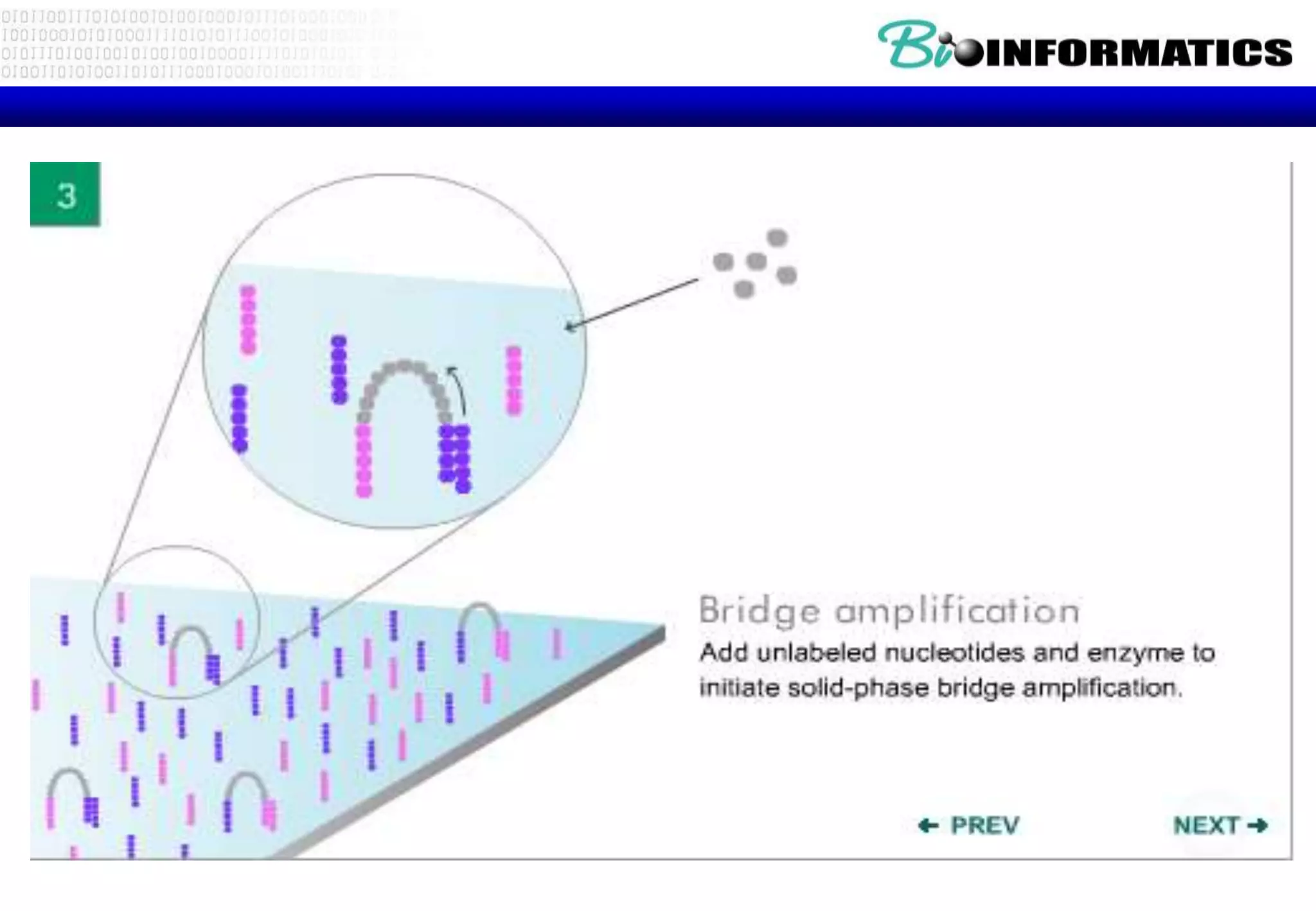
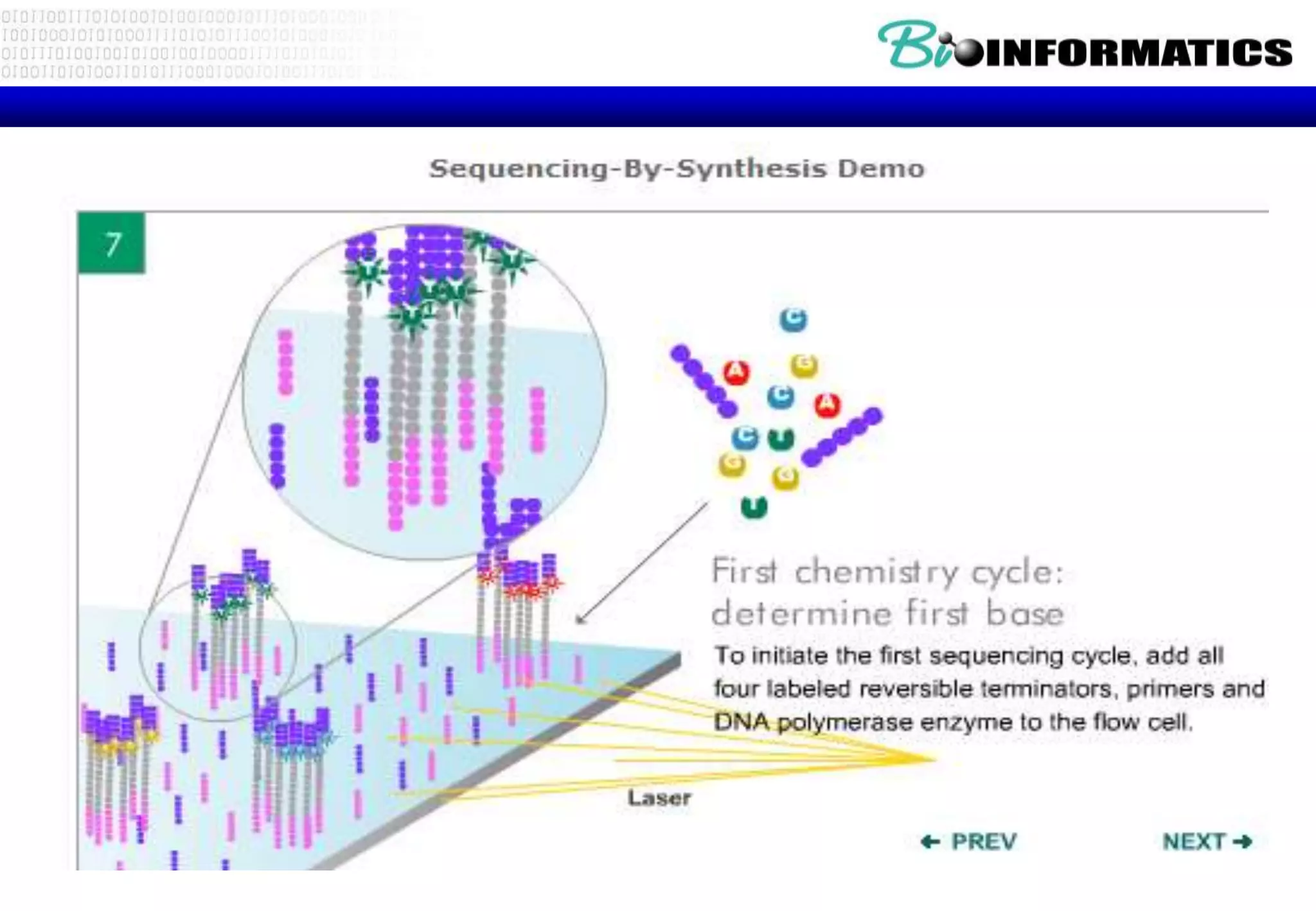
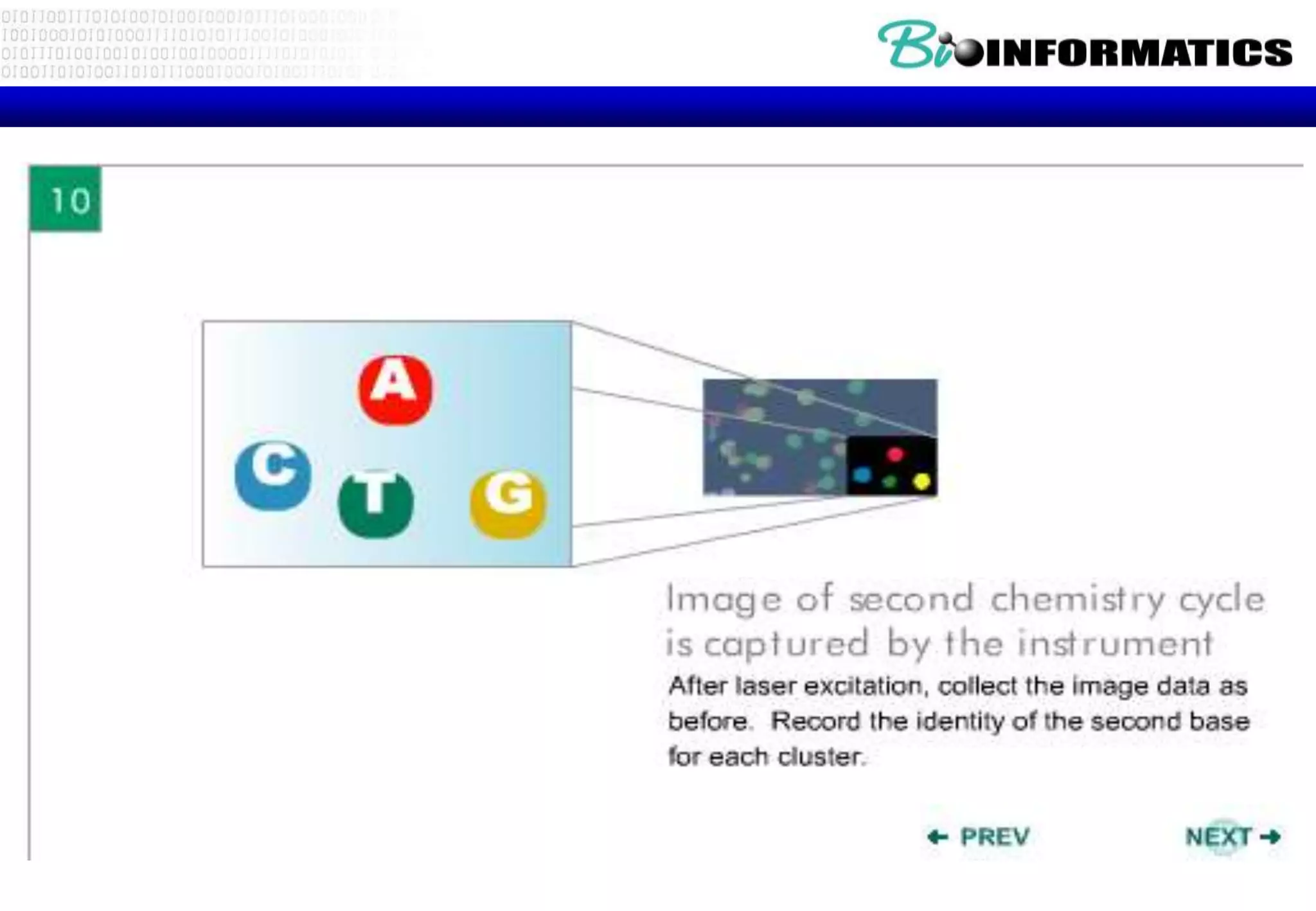
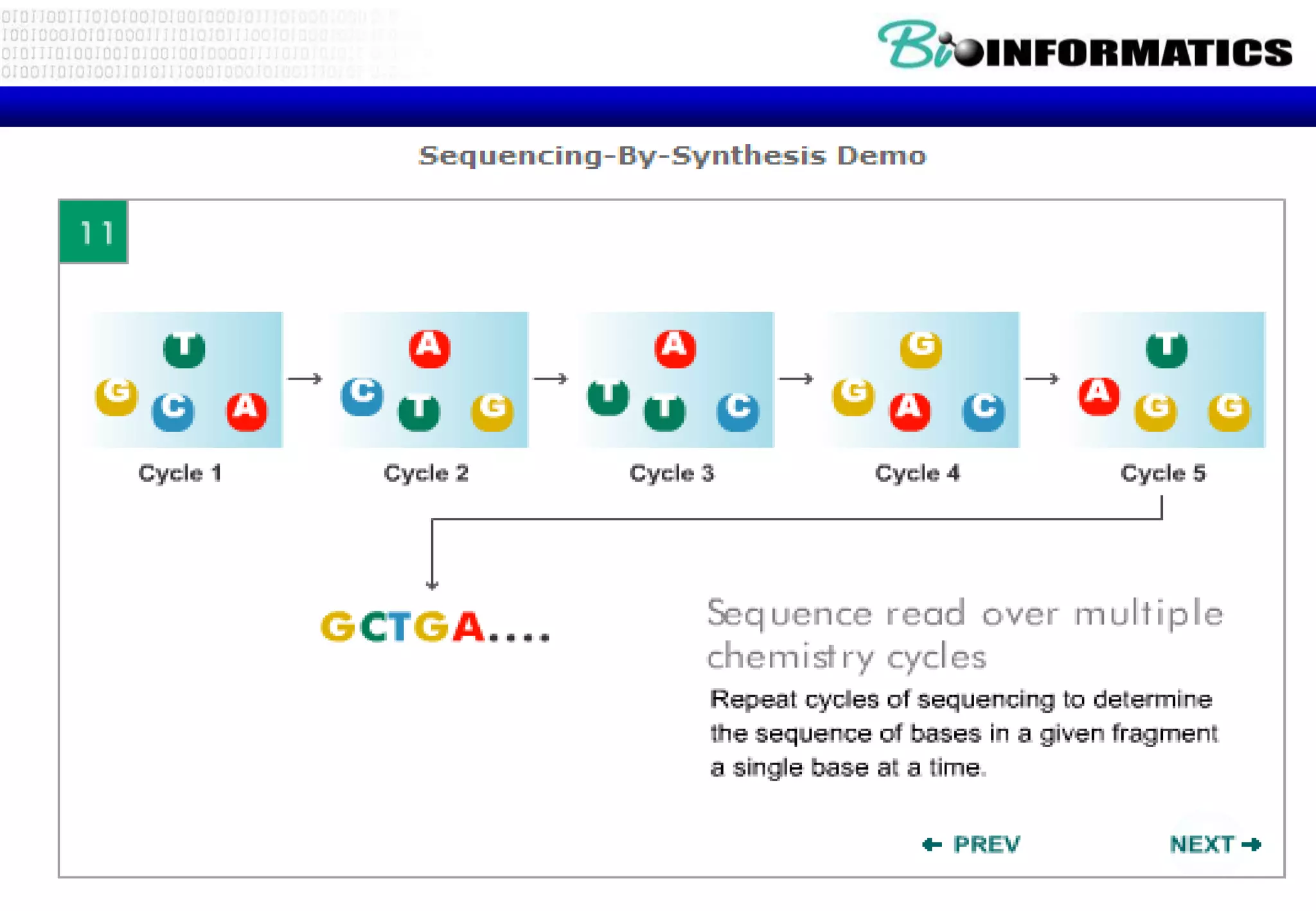
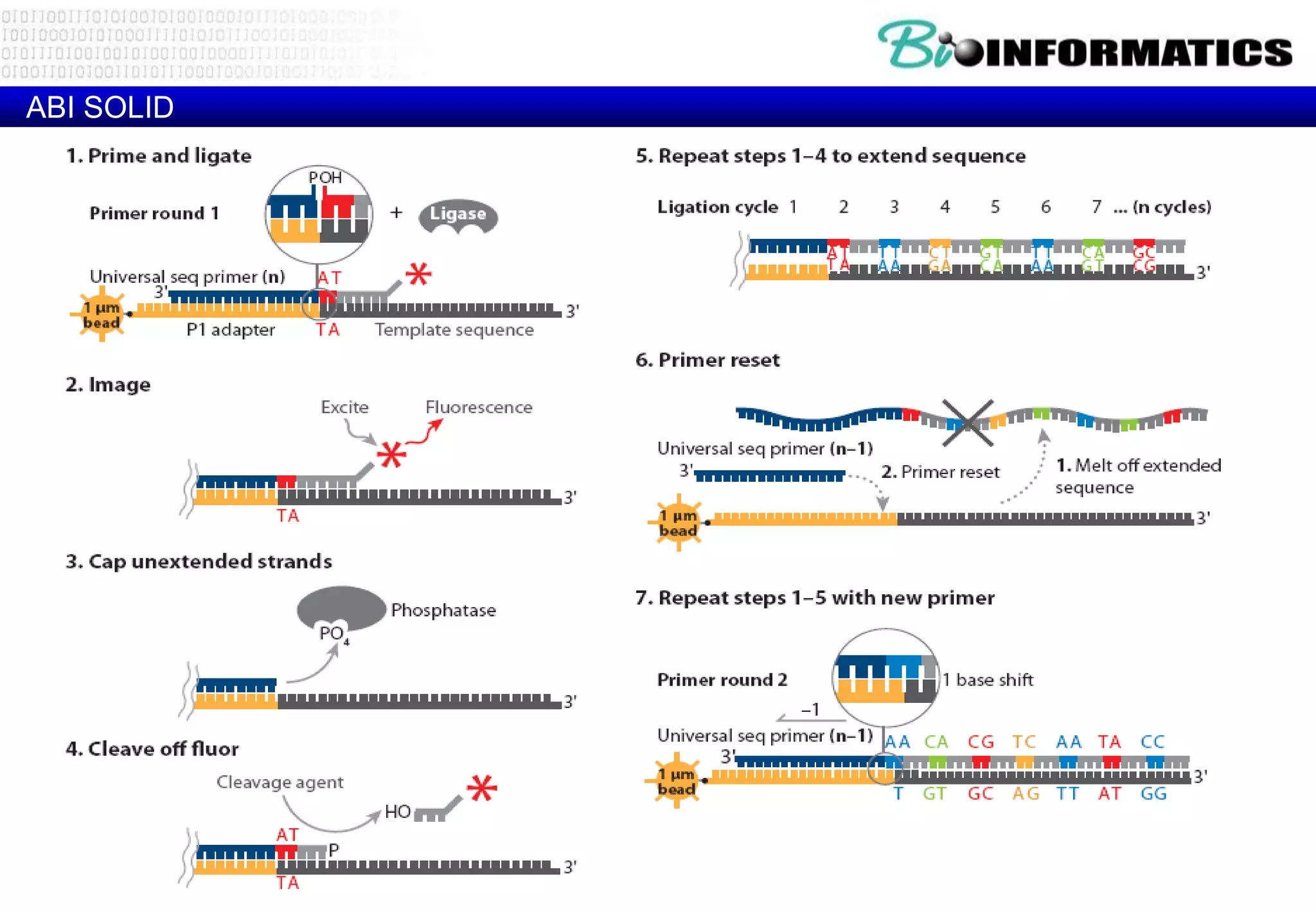
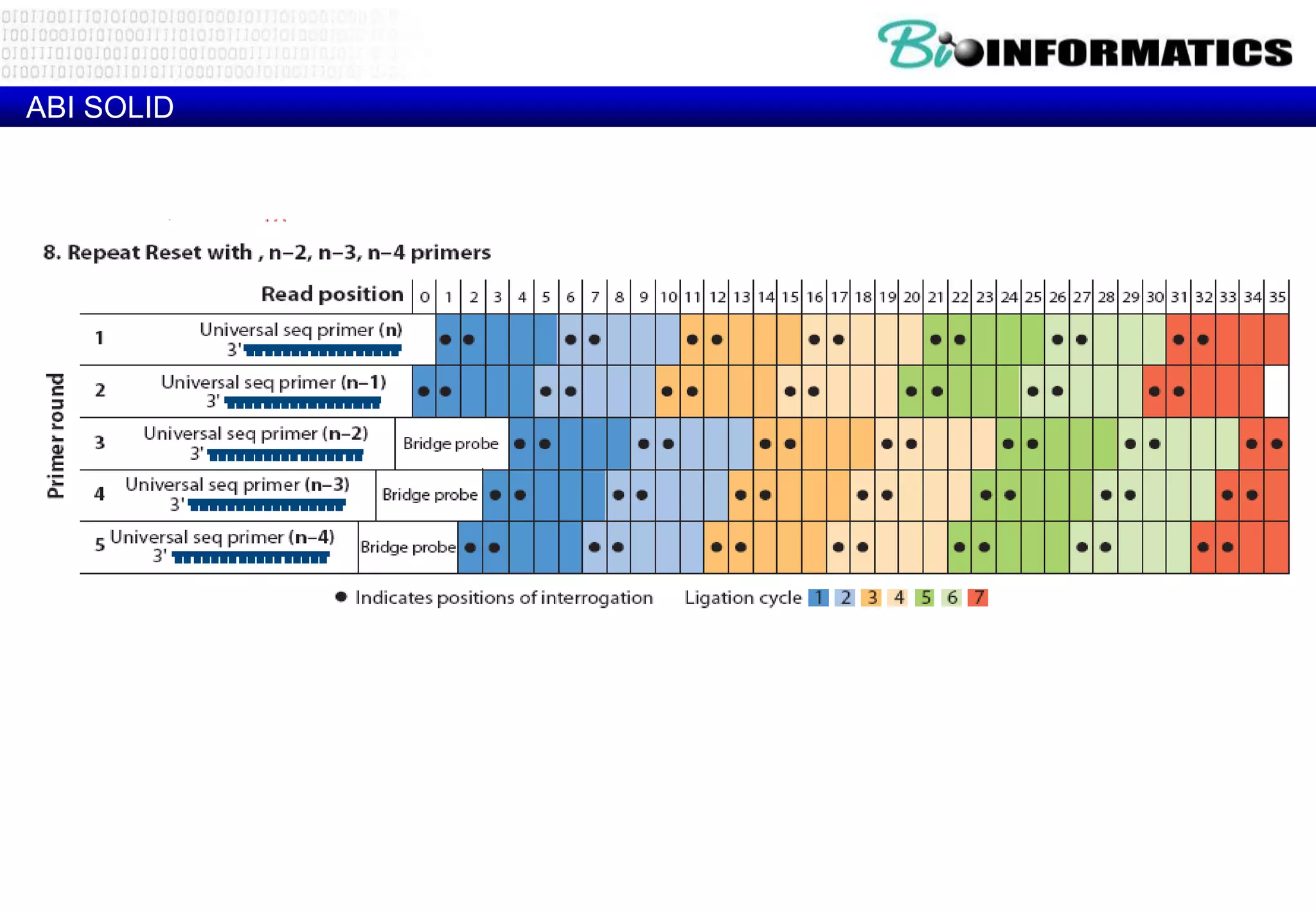
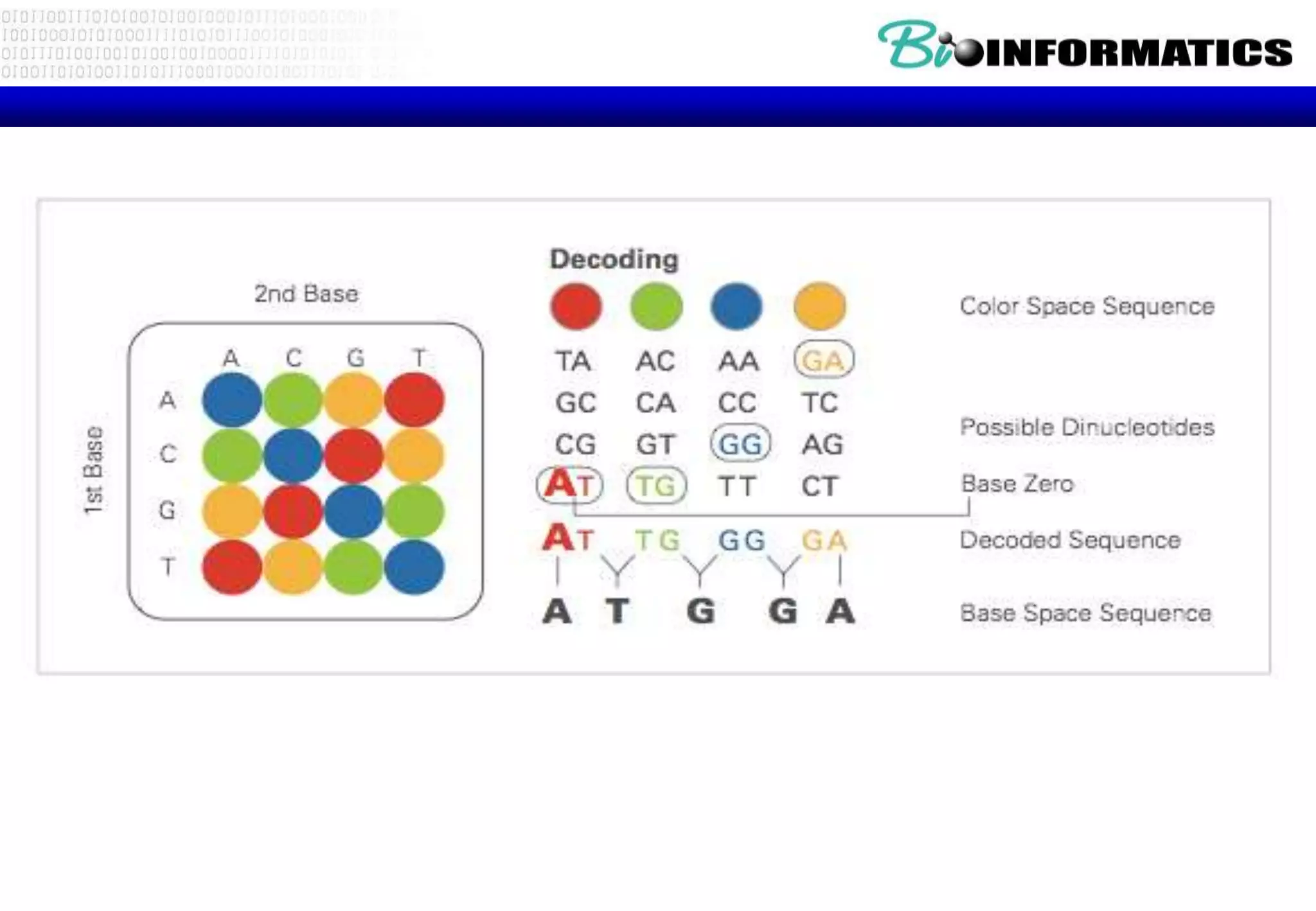
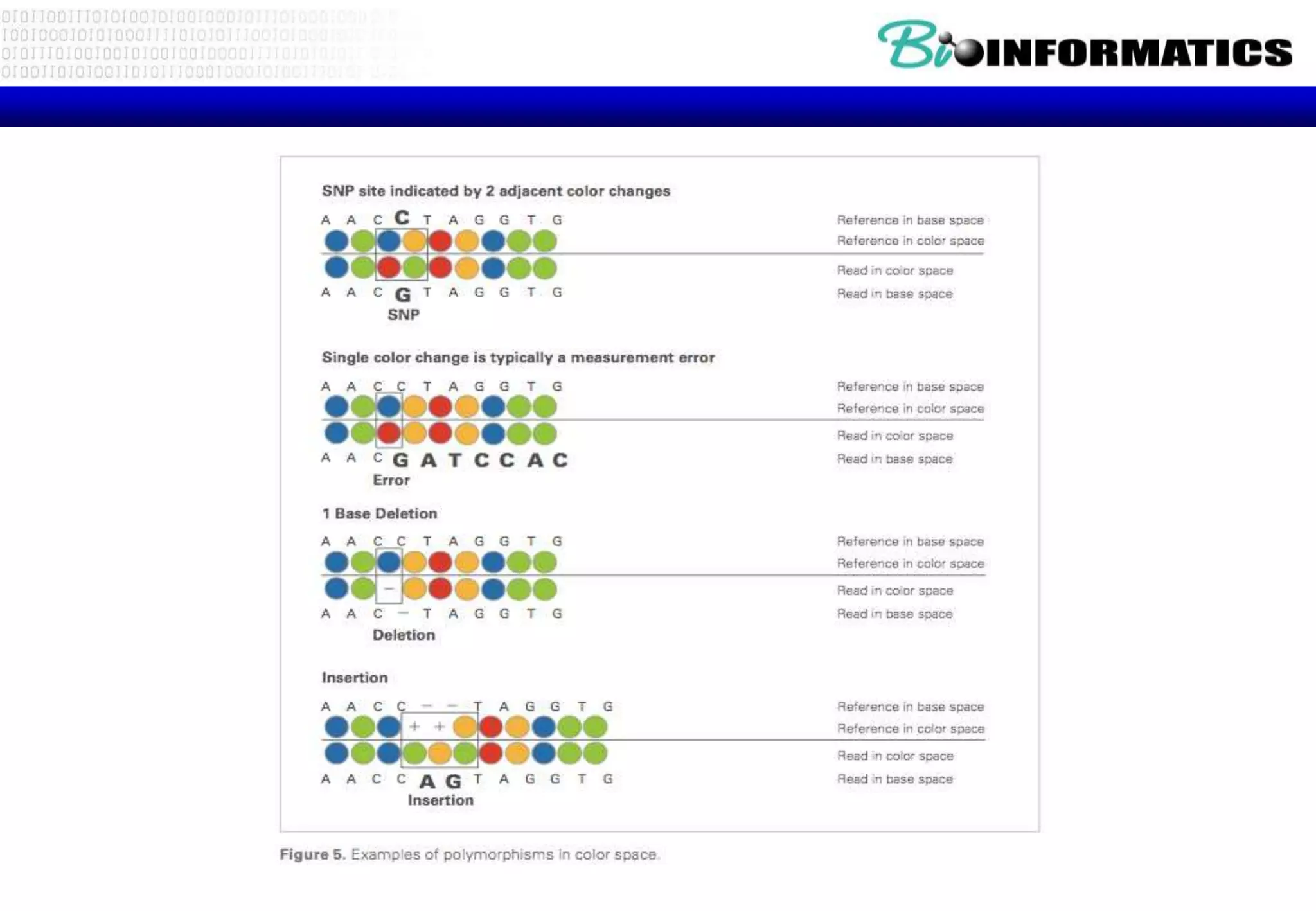
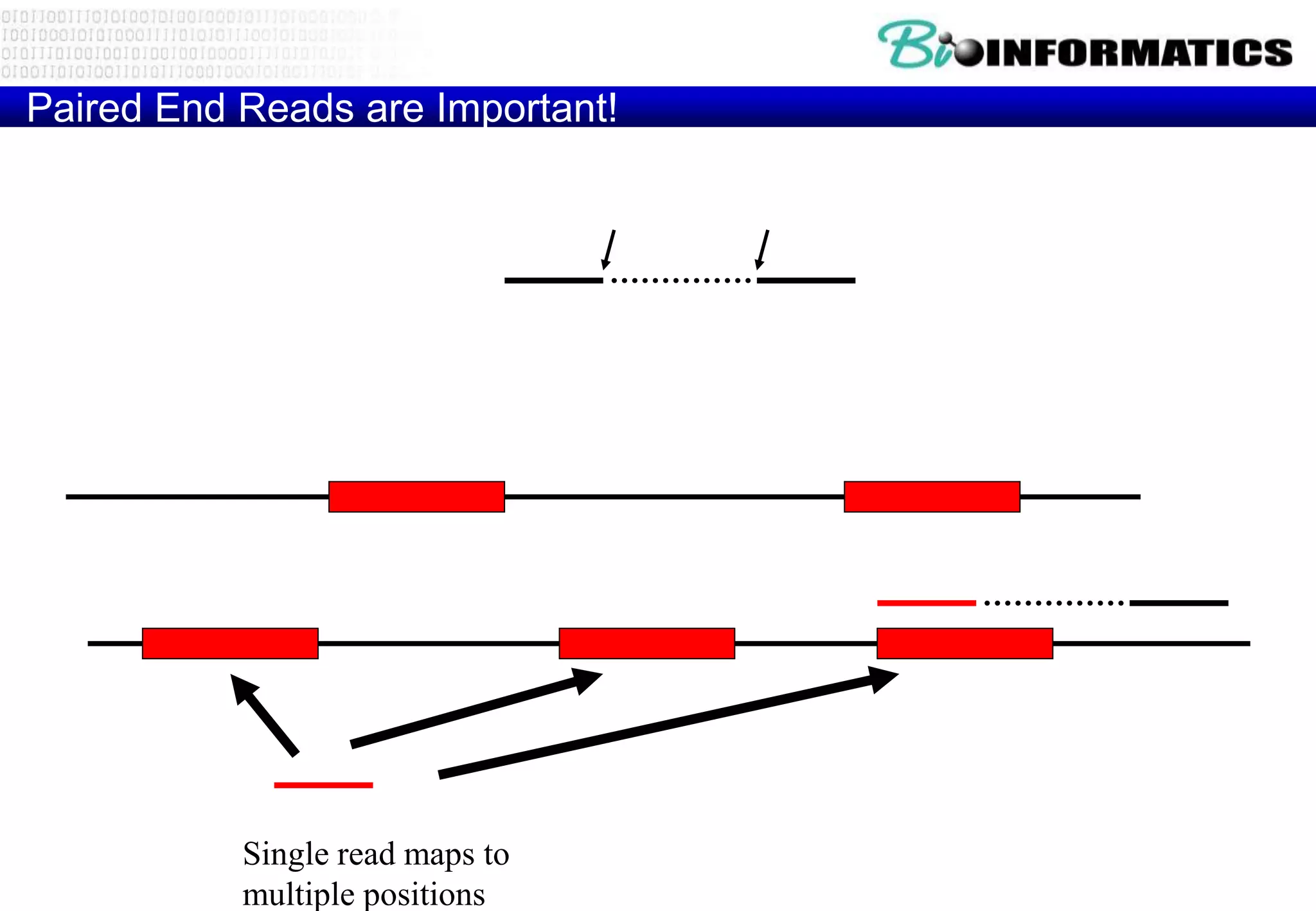
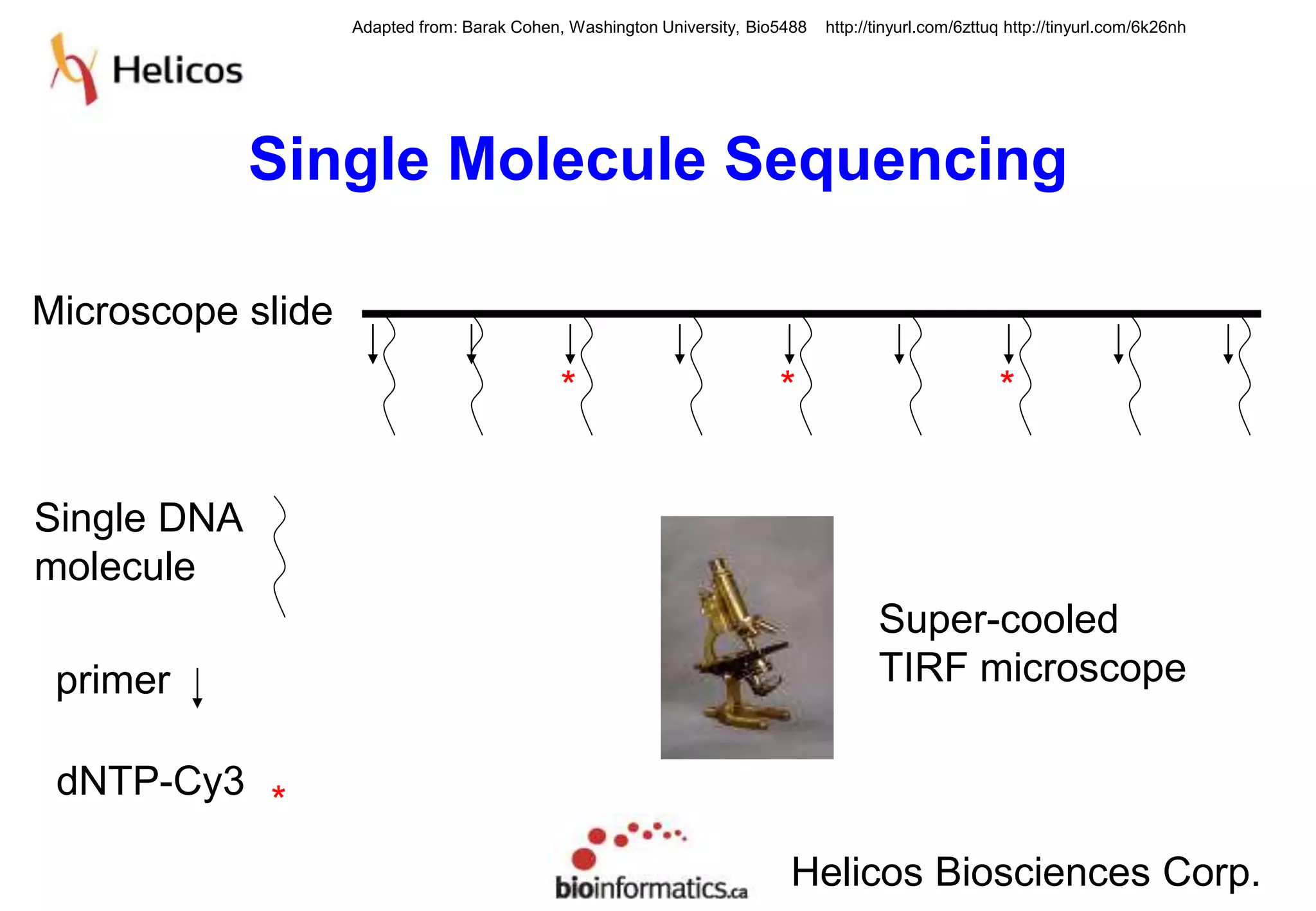
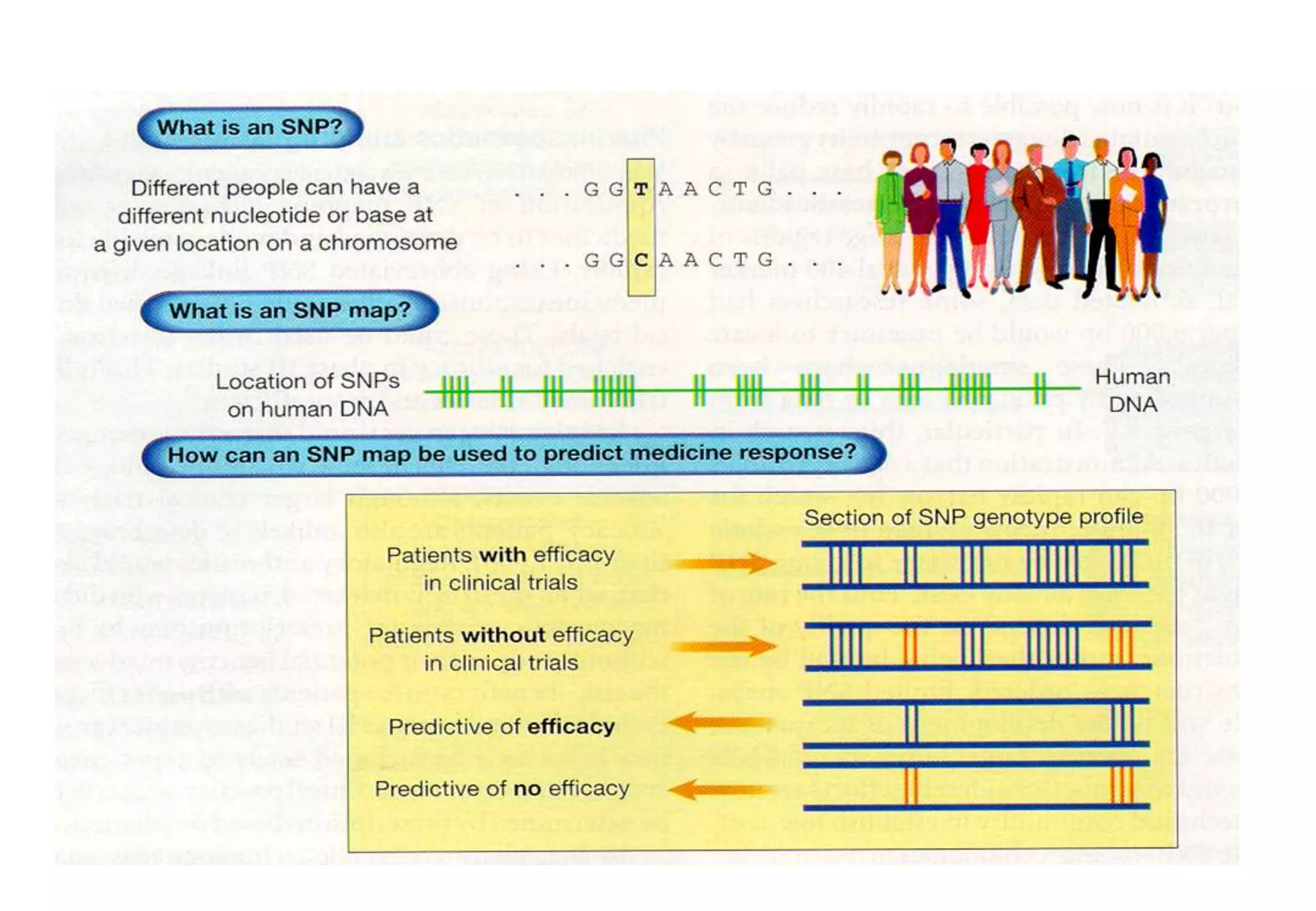
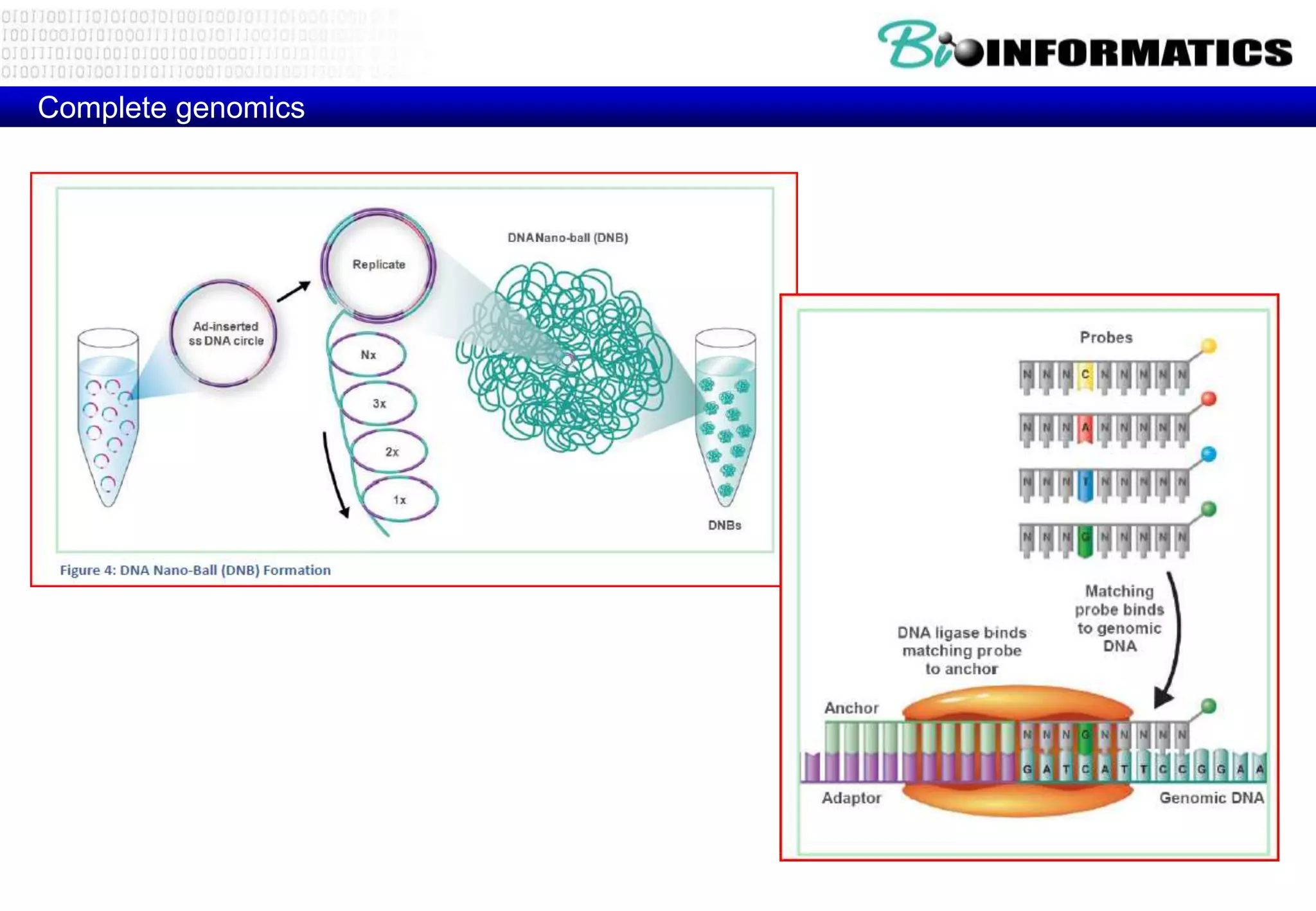
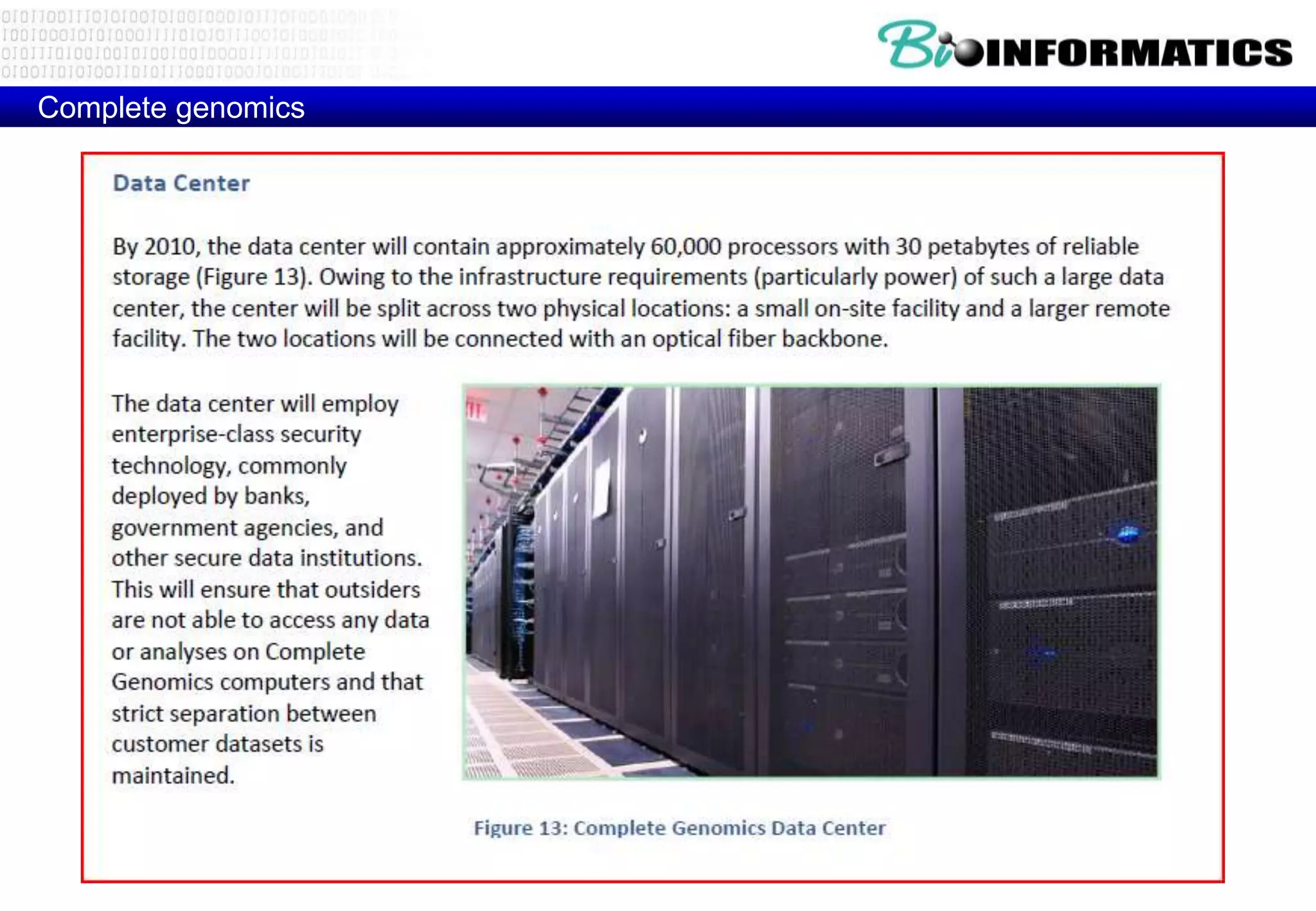
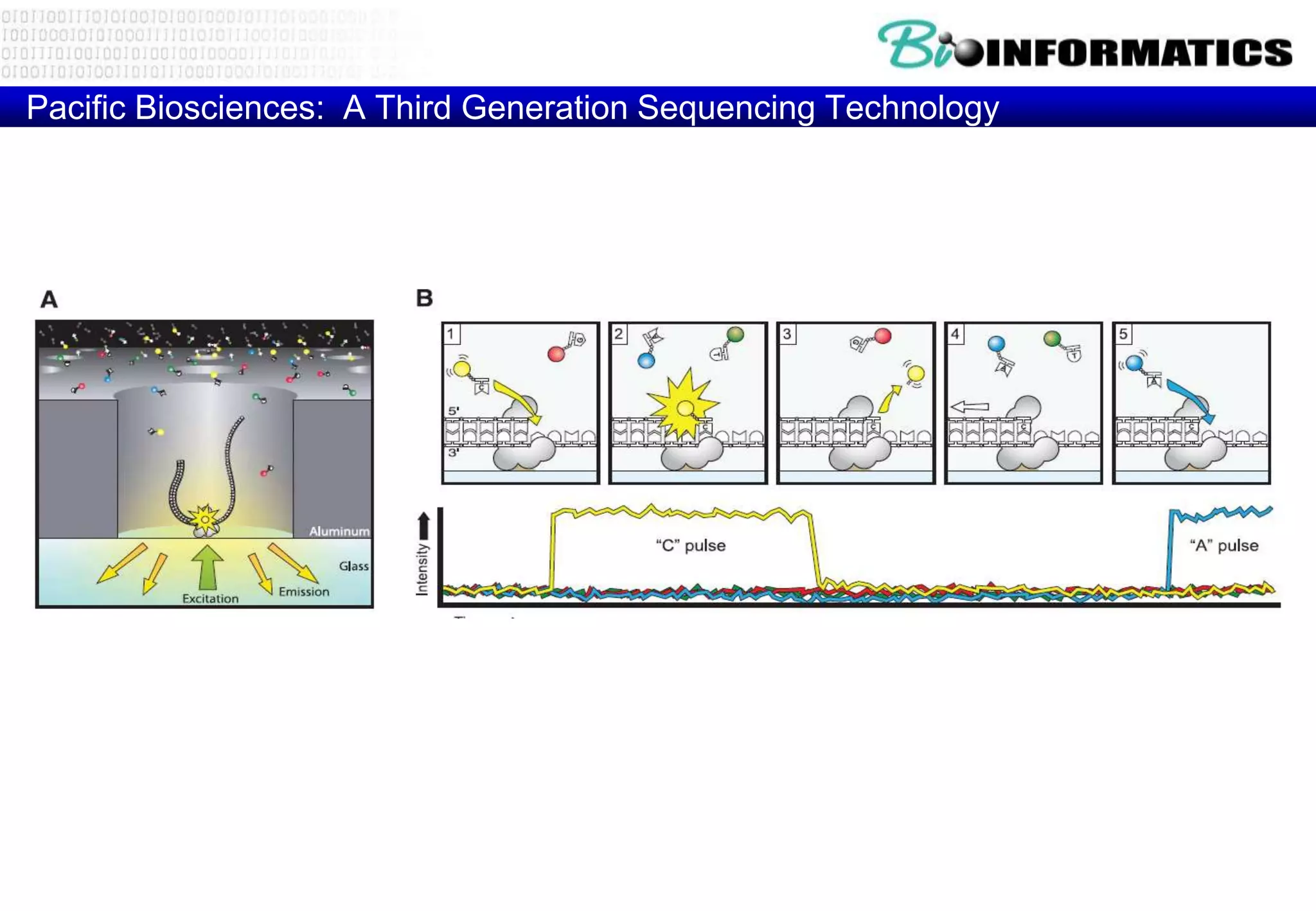
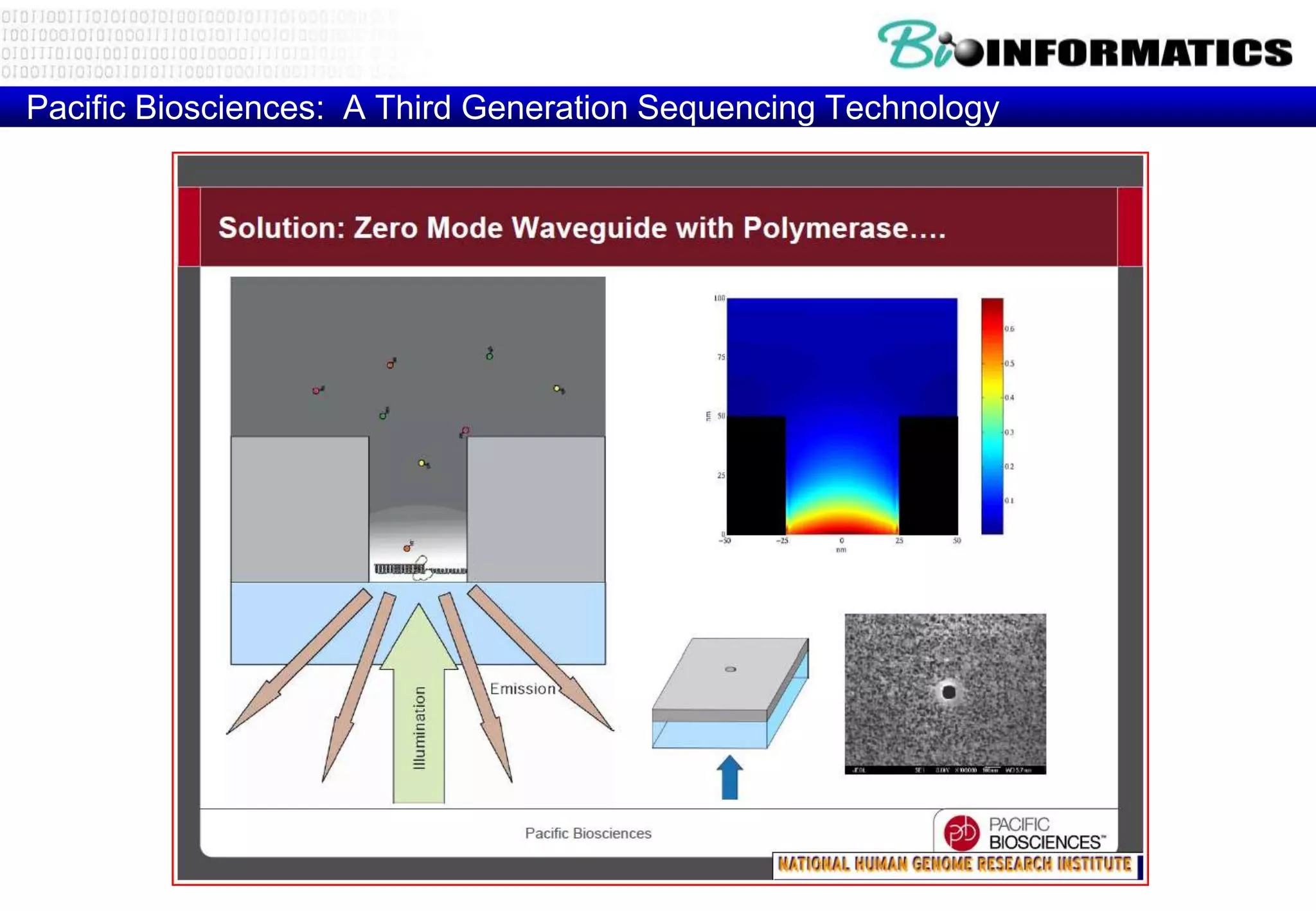
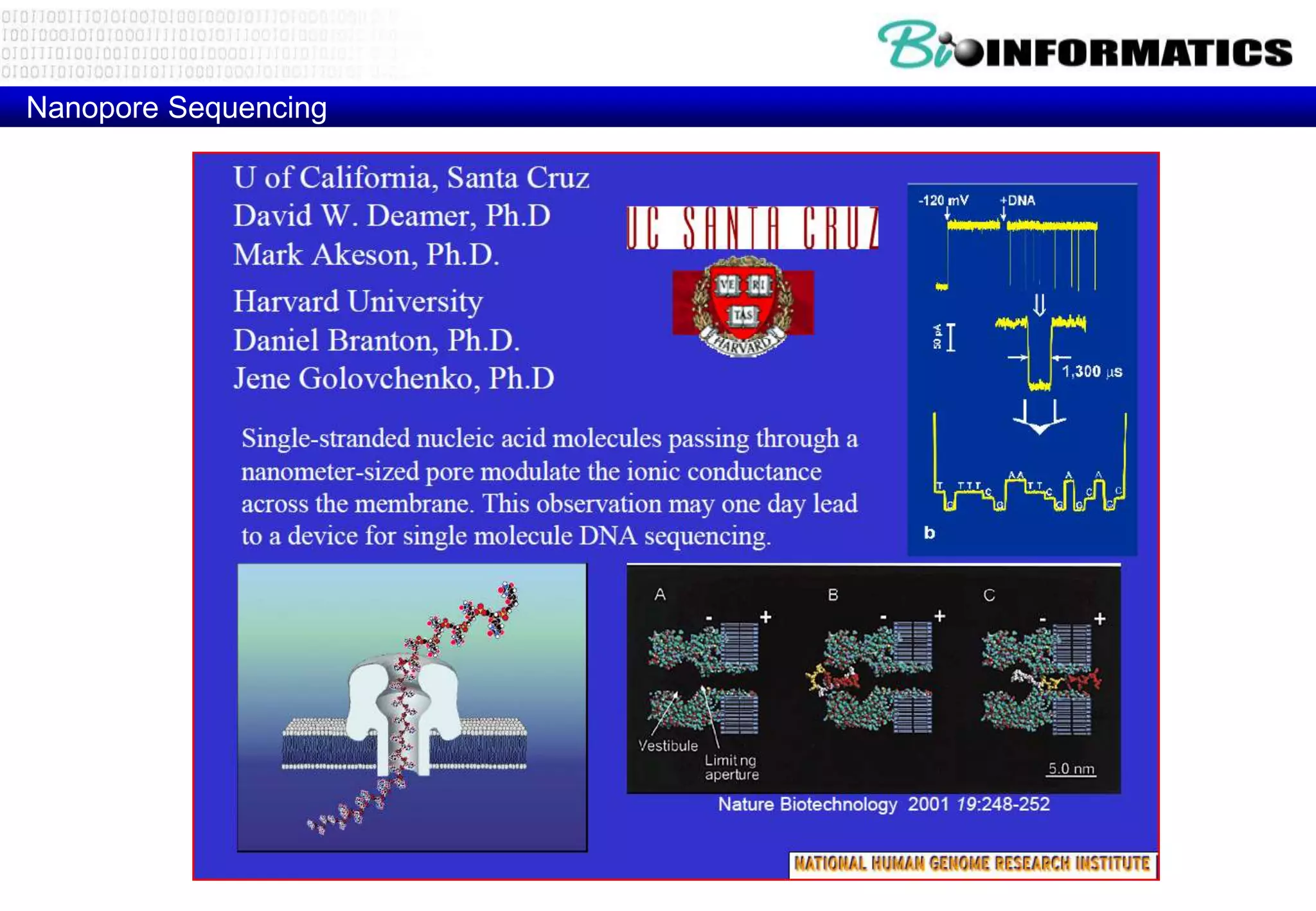
This document provides an overview of bioinformatics and discusses key topics in the field. It begins by defining bioinformatics as the application of information technology to the analysis and management of biological data, facilitated by the use of computers. It then lists some common applications of bioinformatics like sequence analysis, molecular modeling, phylogeny analysis, and medical informatics. The document also discusses some of the promises of genomics and bioinformatics for applications in medicine, agriculture, and other fields. It provides a brief history of the emergence of bioinformatics as a field in the 1990s. Finally, it outlines some of the main topics that will be covered in the bioinformatics course, including databases, algorithms, interface design, and computational methods.