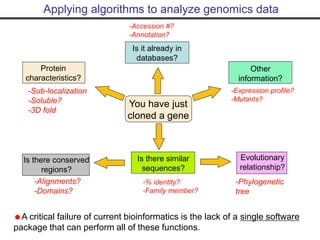
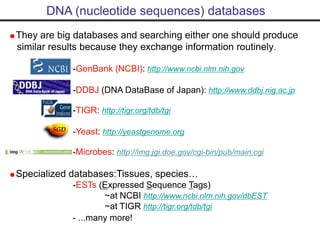
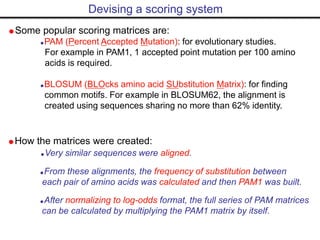
This document provides an overview of bioinformatics and discusses key concepts like:
- Bioinformatics combines biology, computer science, and information technology to analyze large amounts of biological data.
- High-throughput DNA sequencing has generated vast genomic data that requires bioinformatics tools and databases accessible via the internet to analyze and share.
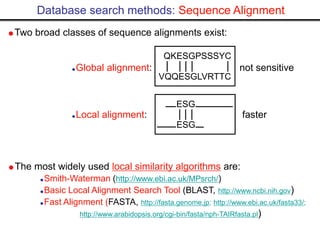
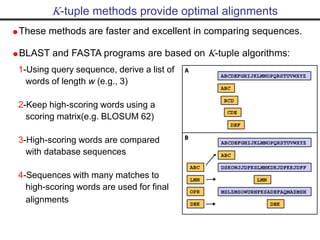
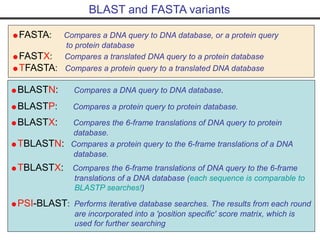
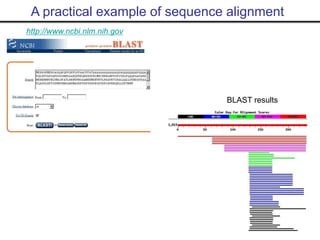
- Popular sequence alignment tools like BLAST, FASTA, and ClustalW are used to search databases and compare sequences, helping researchers analyze genes and genomes.