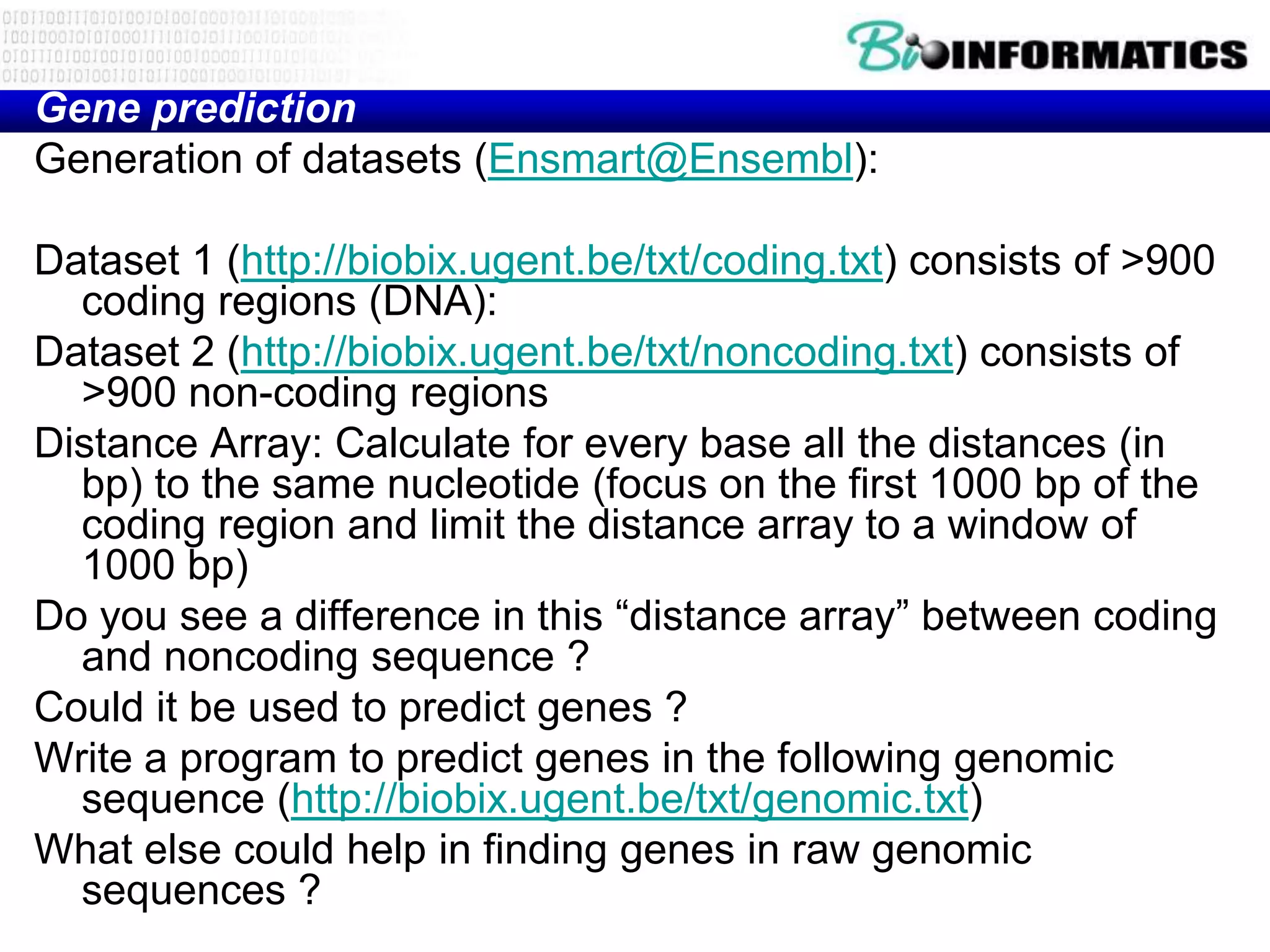
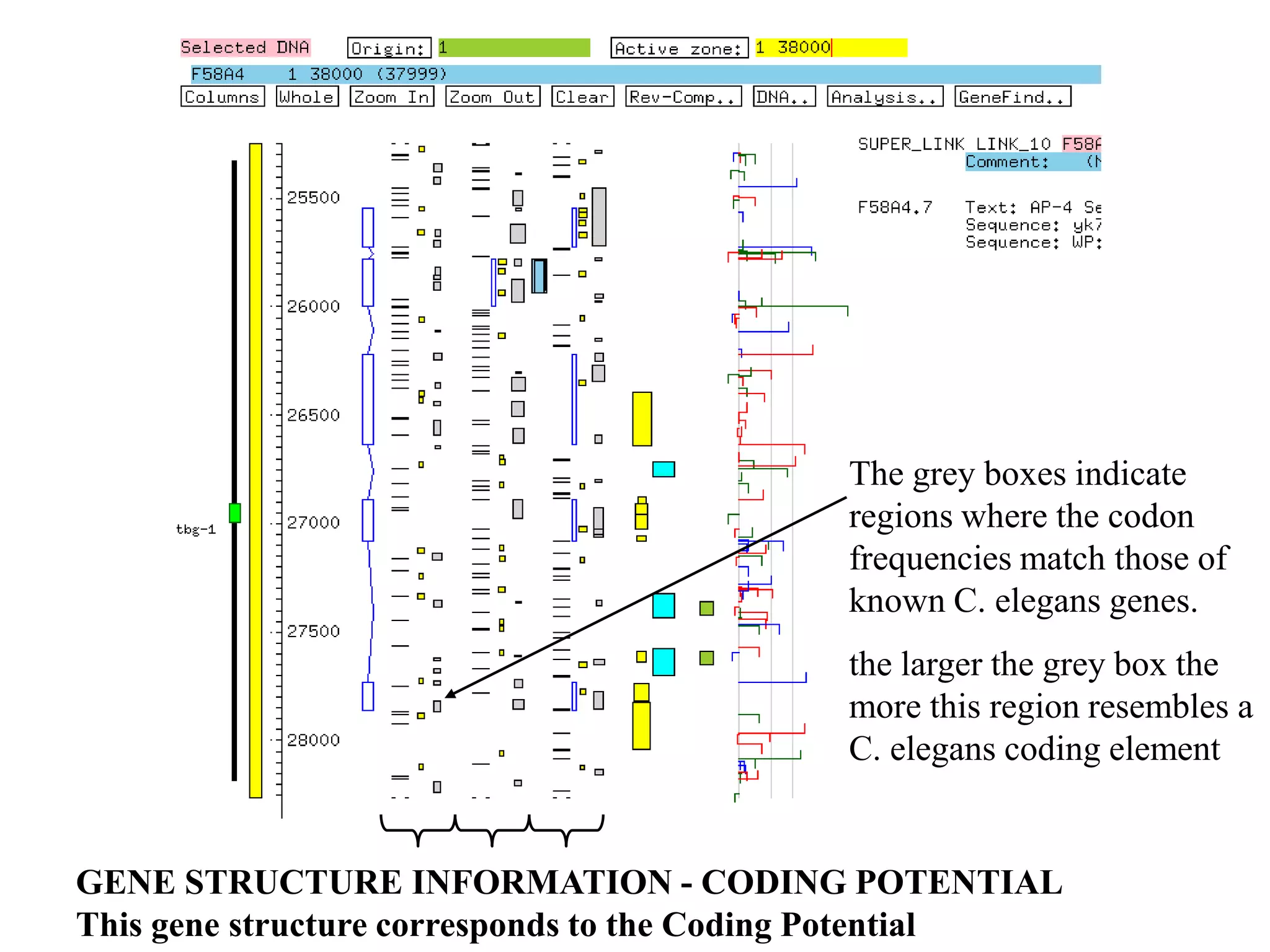
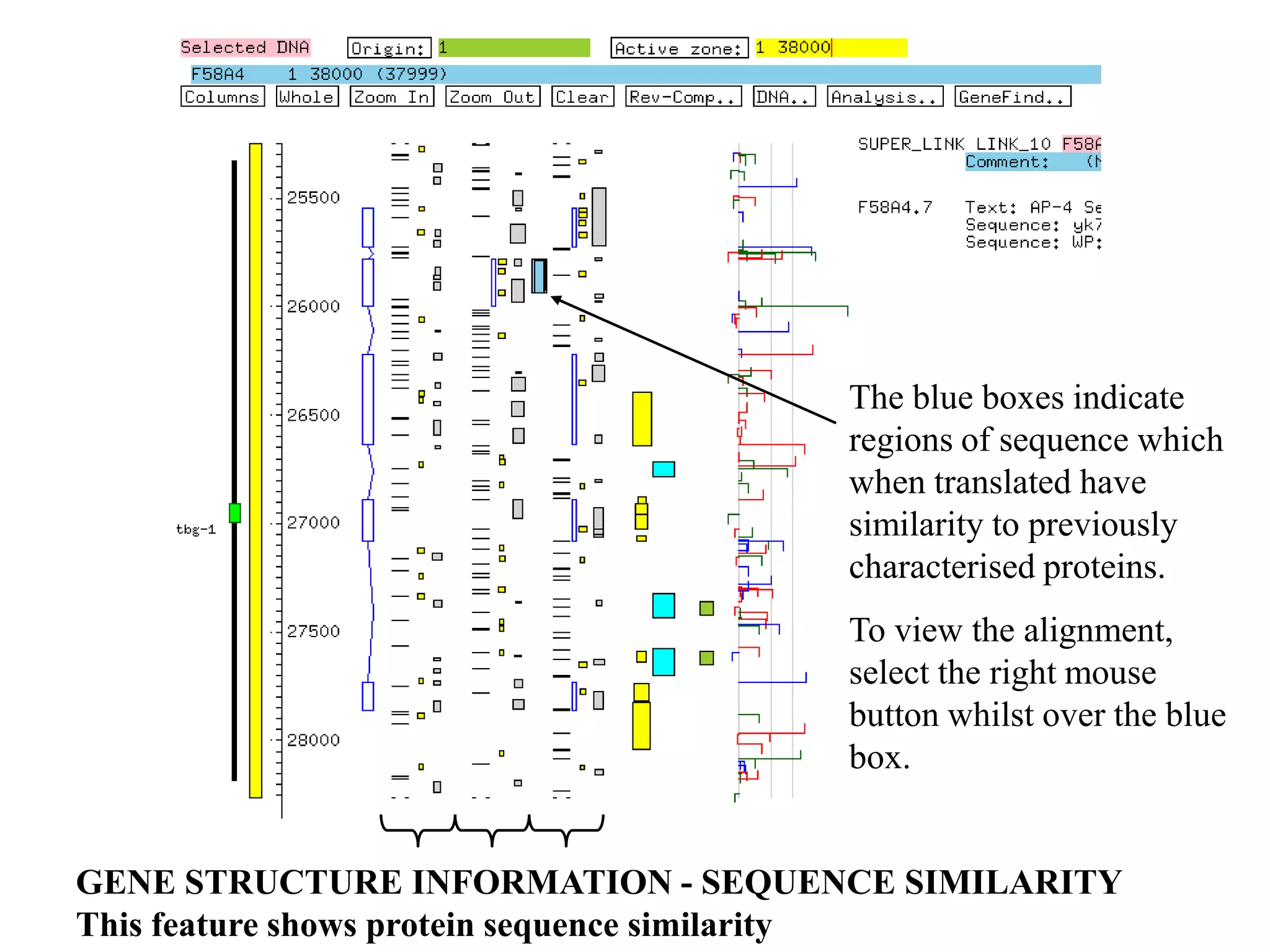
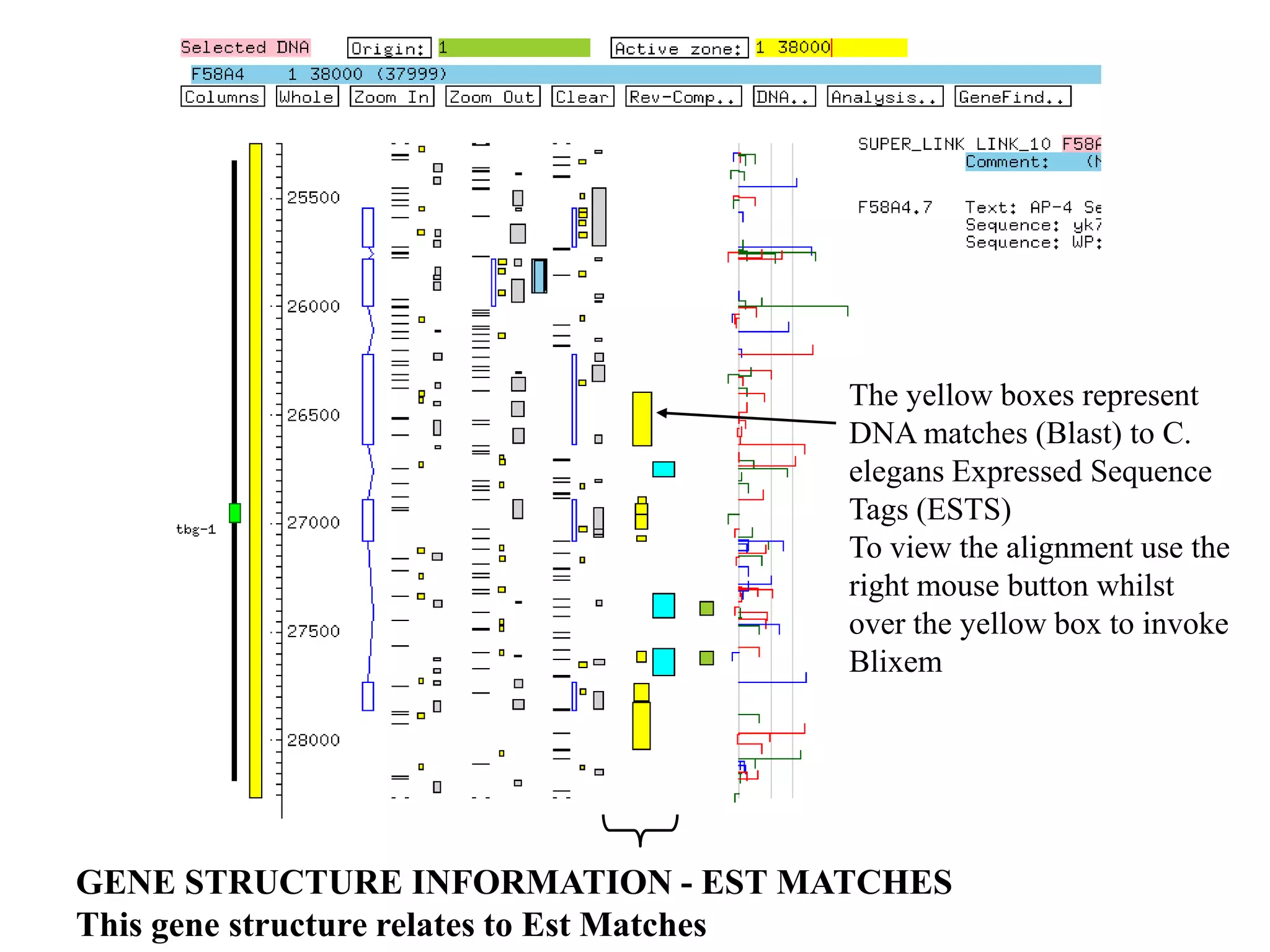
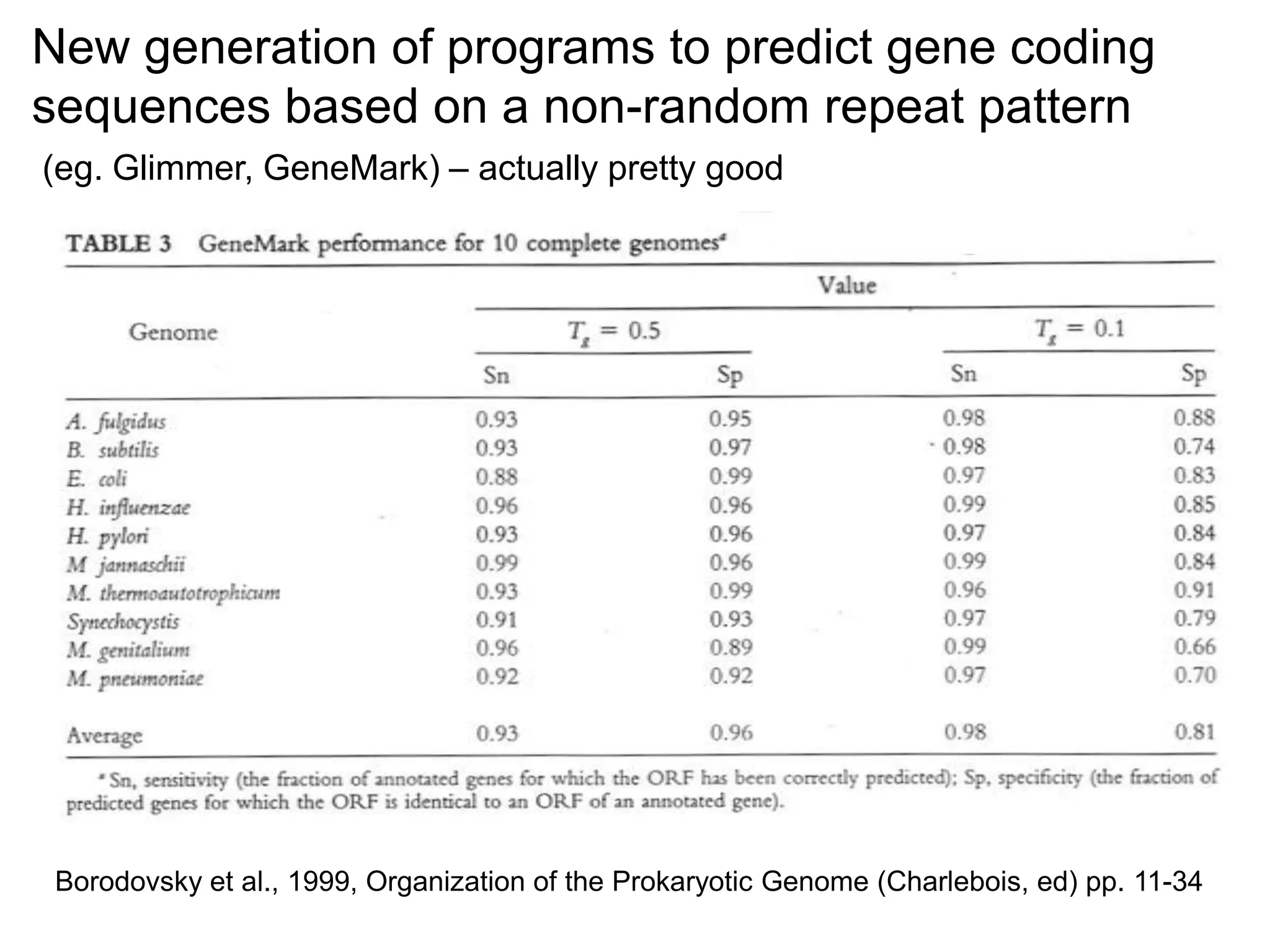
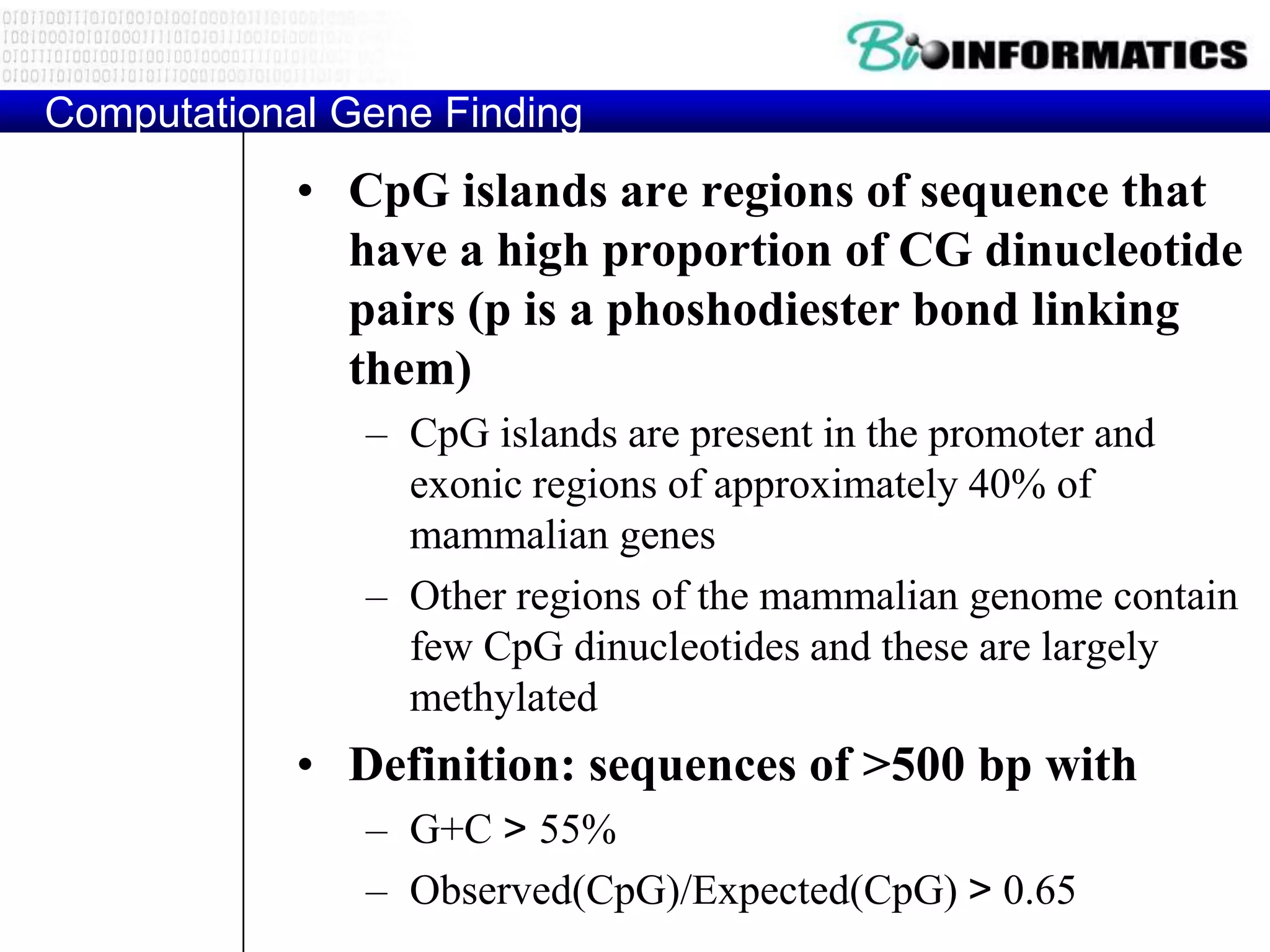
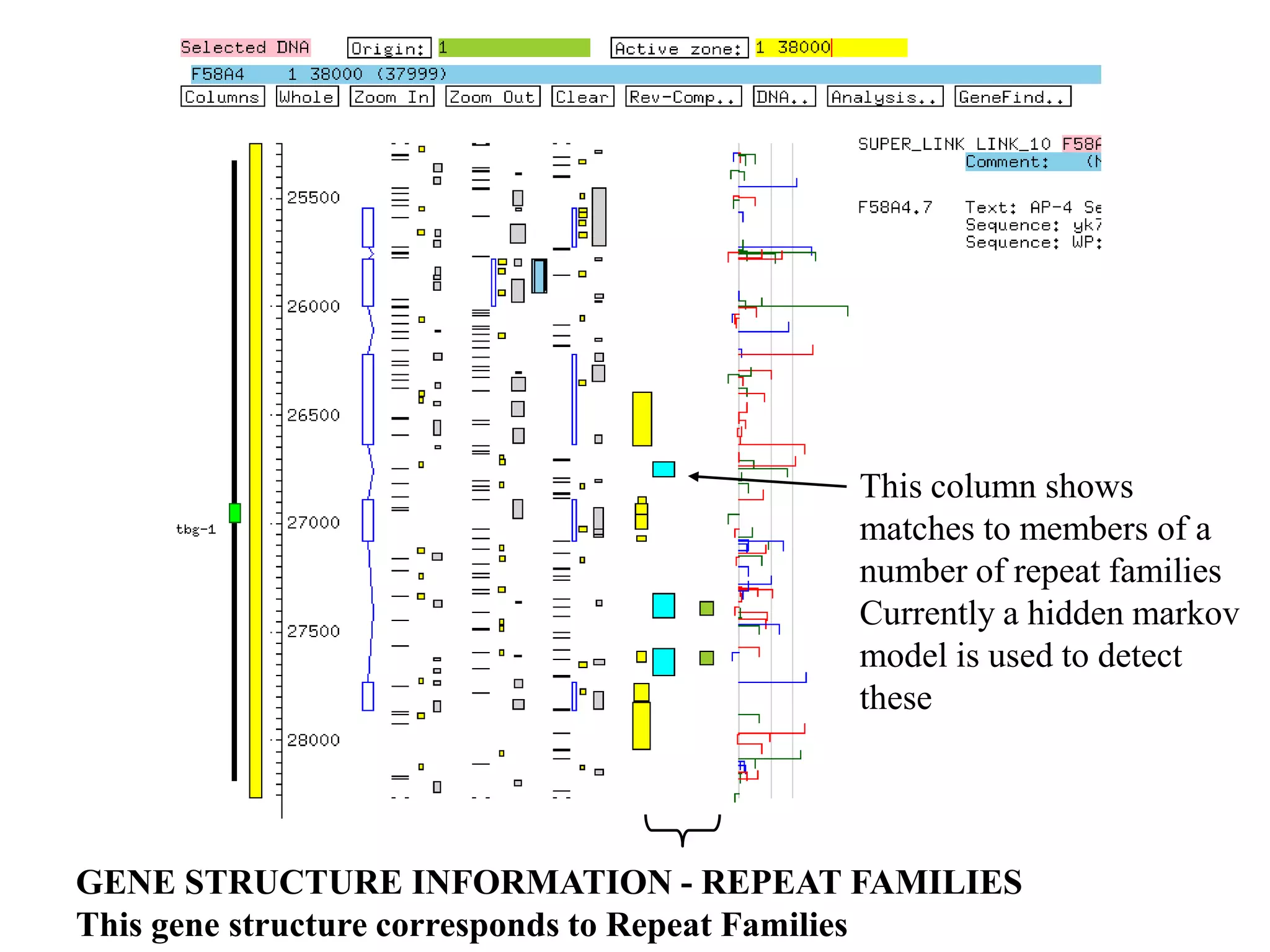
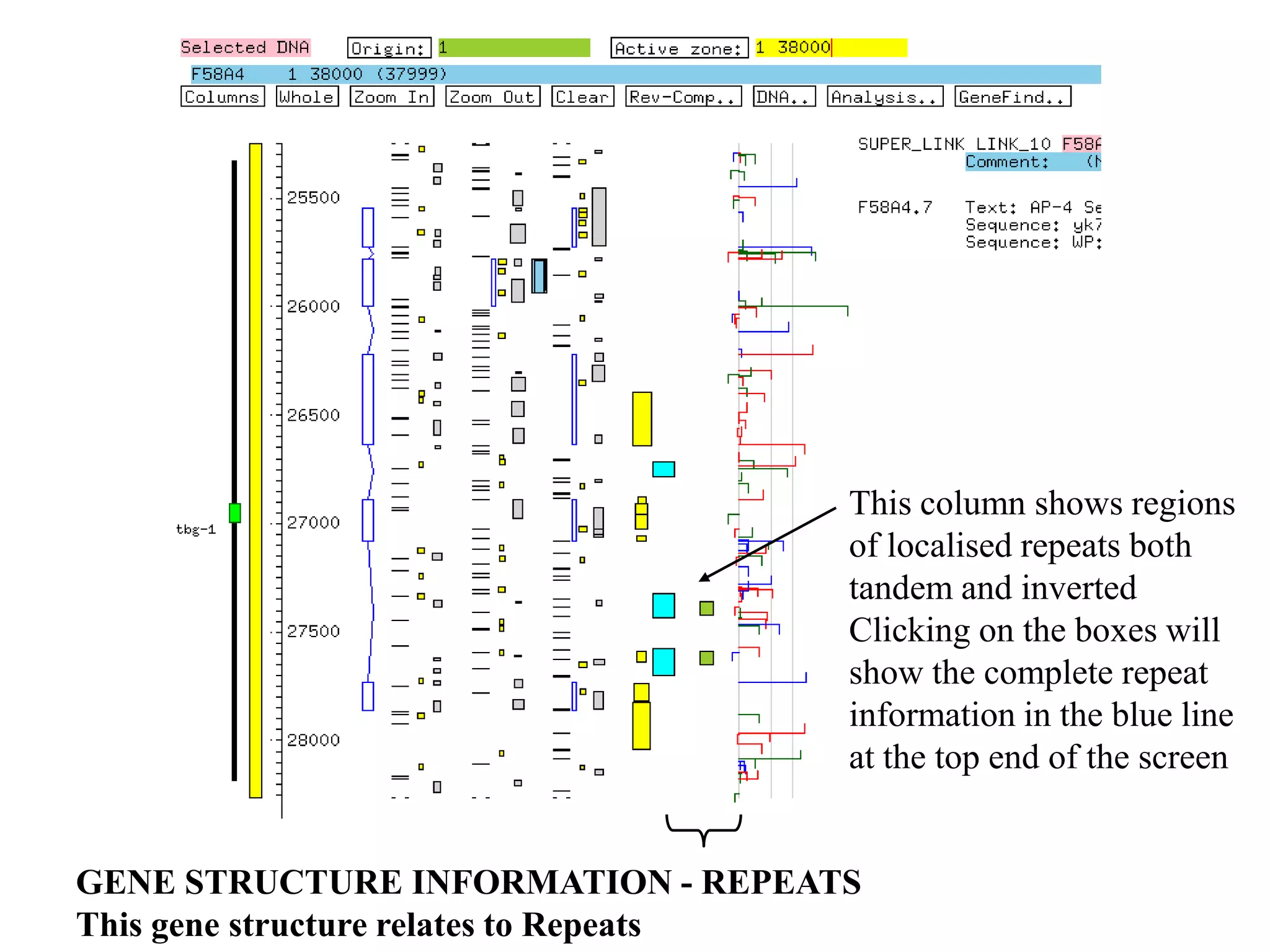
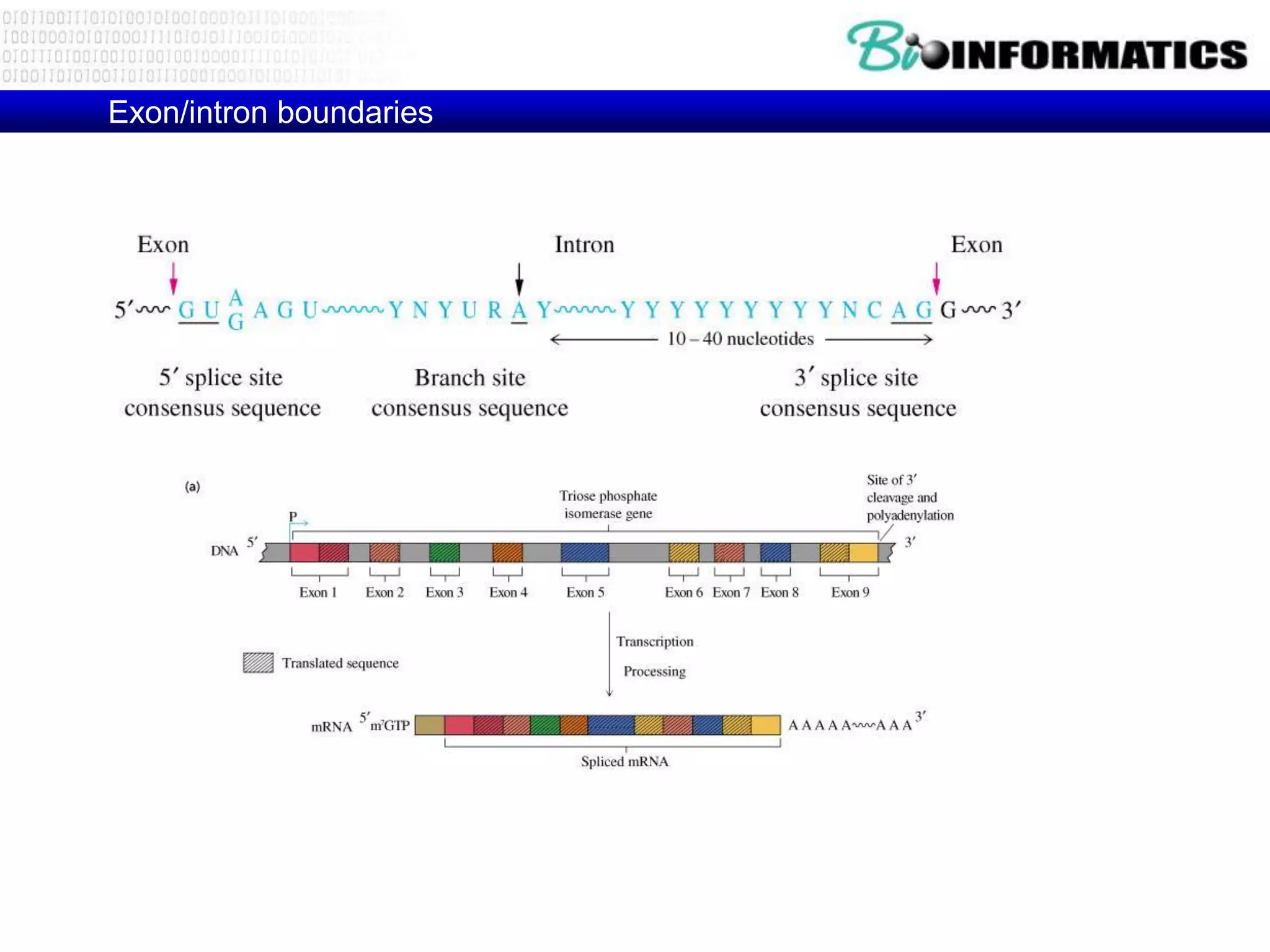
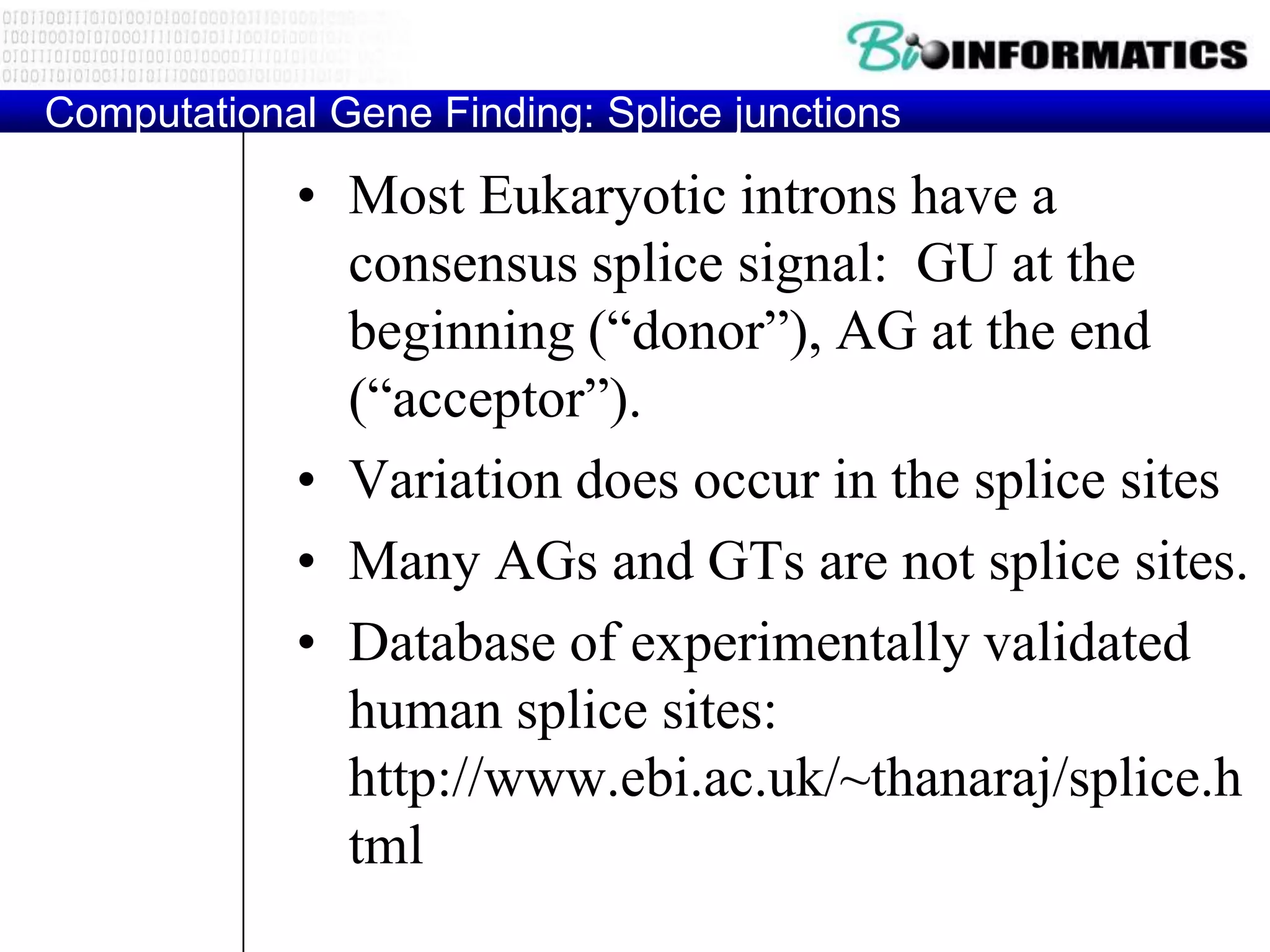
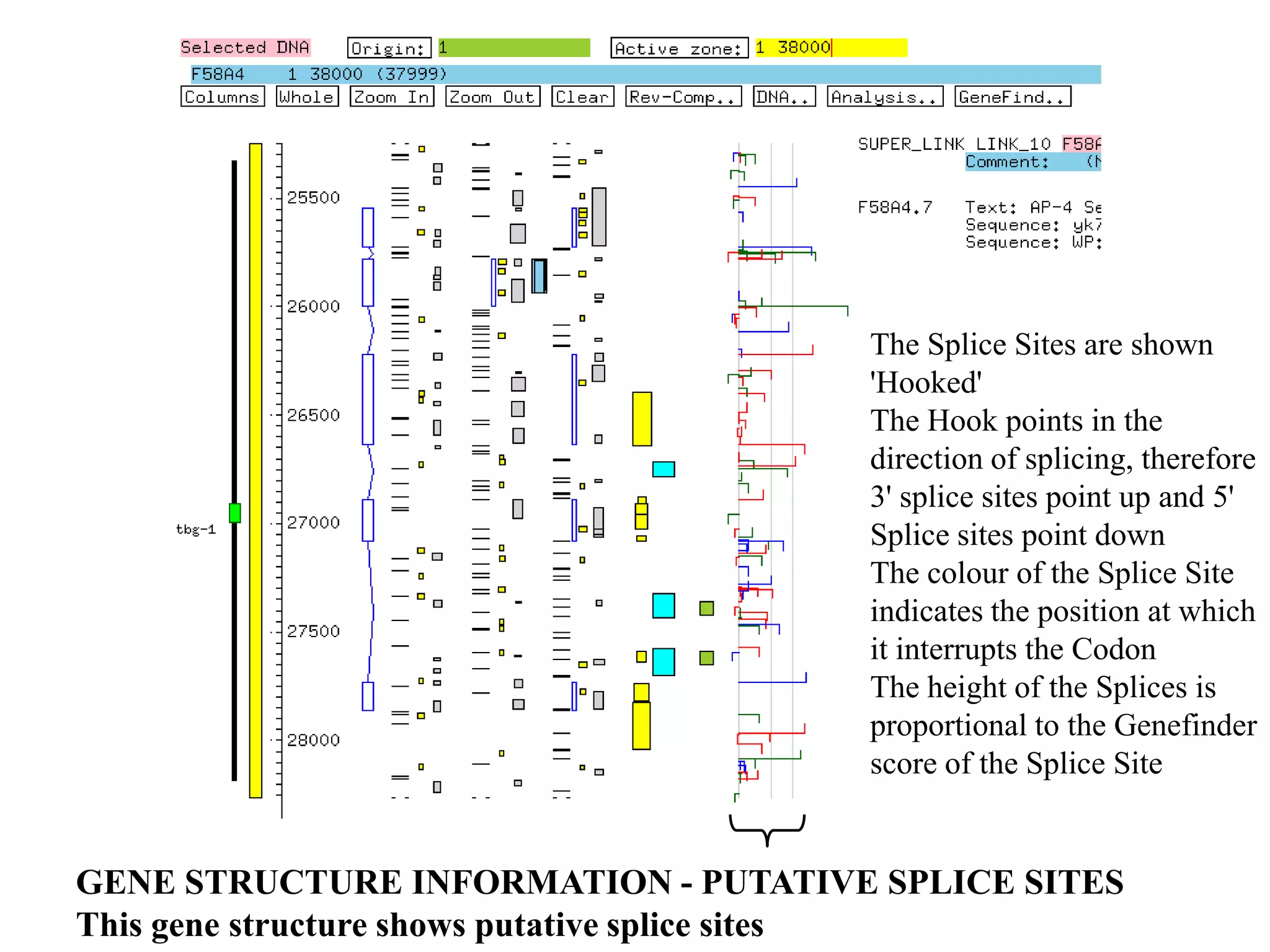
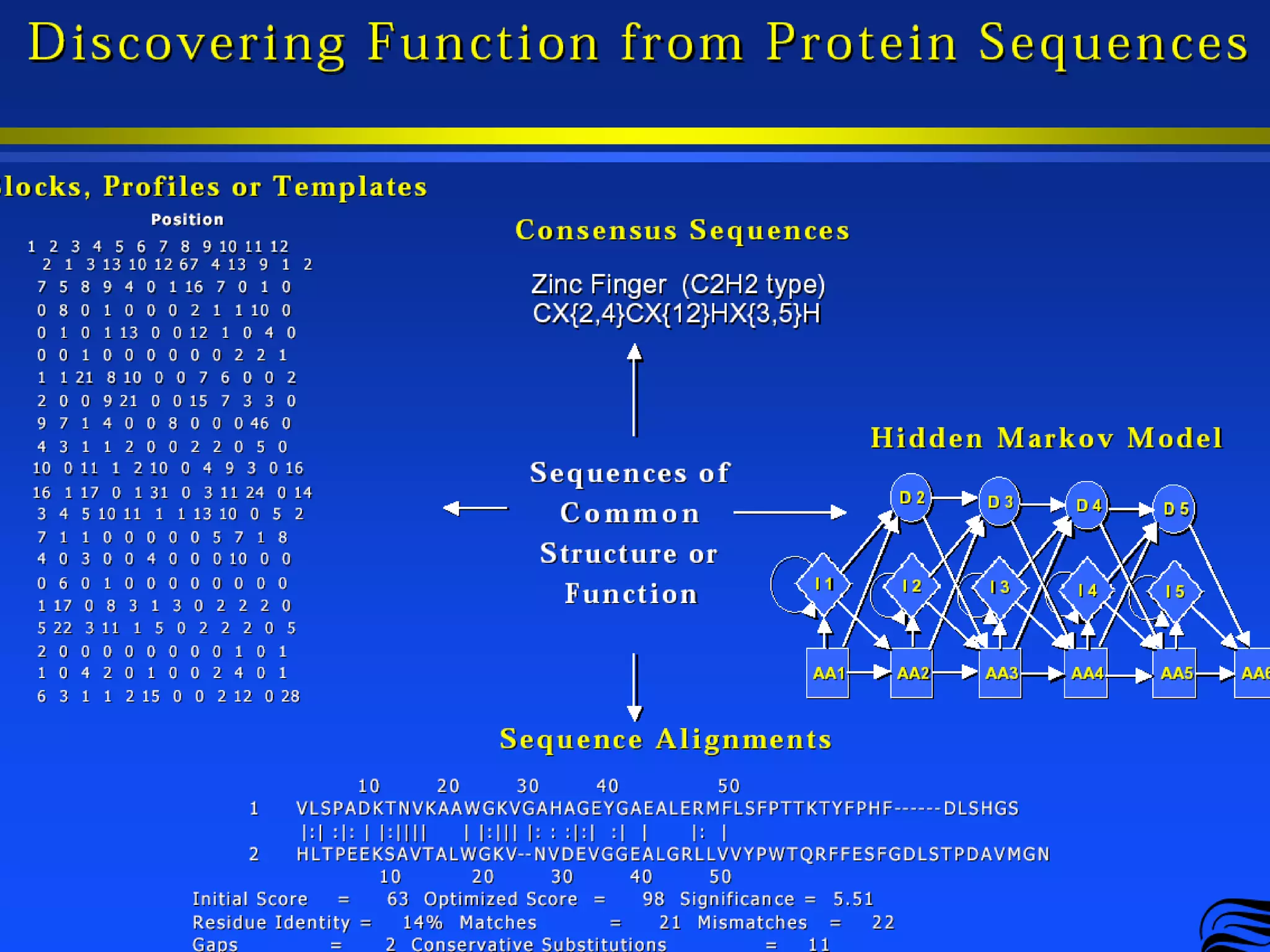
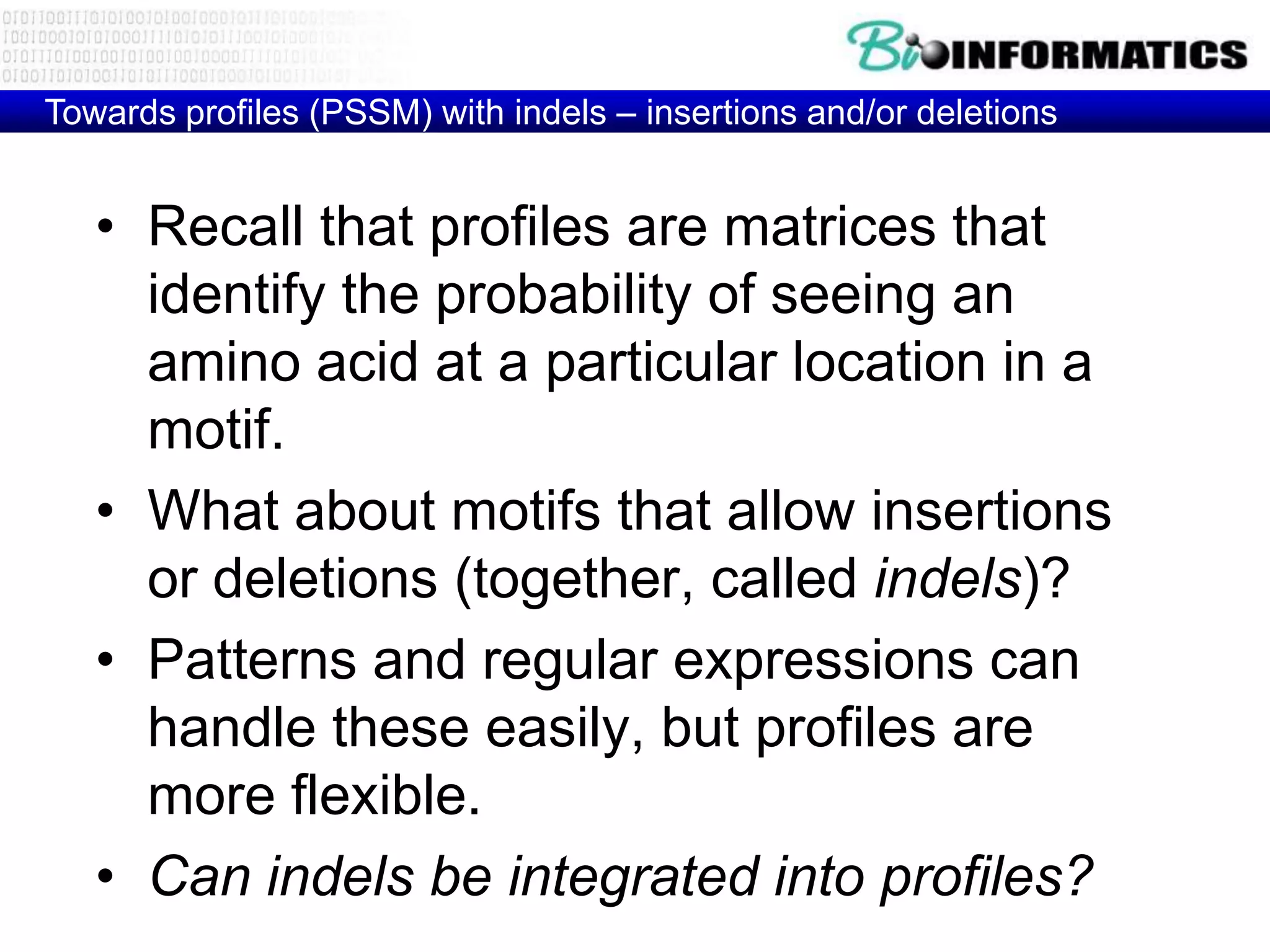
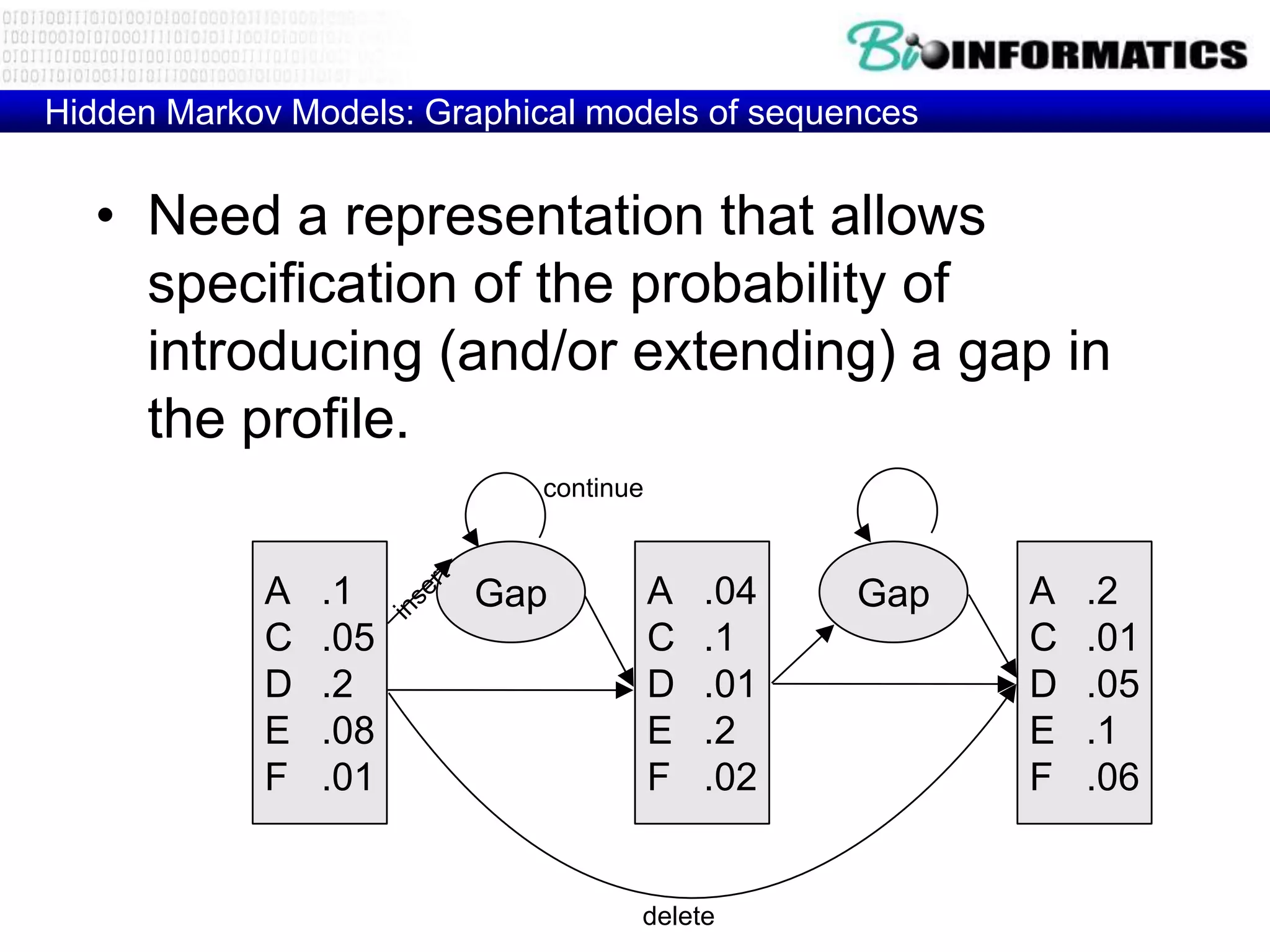
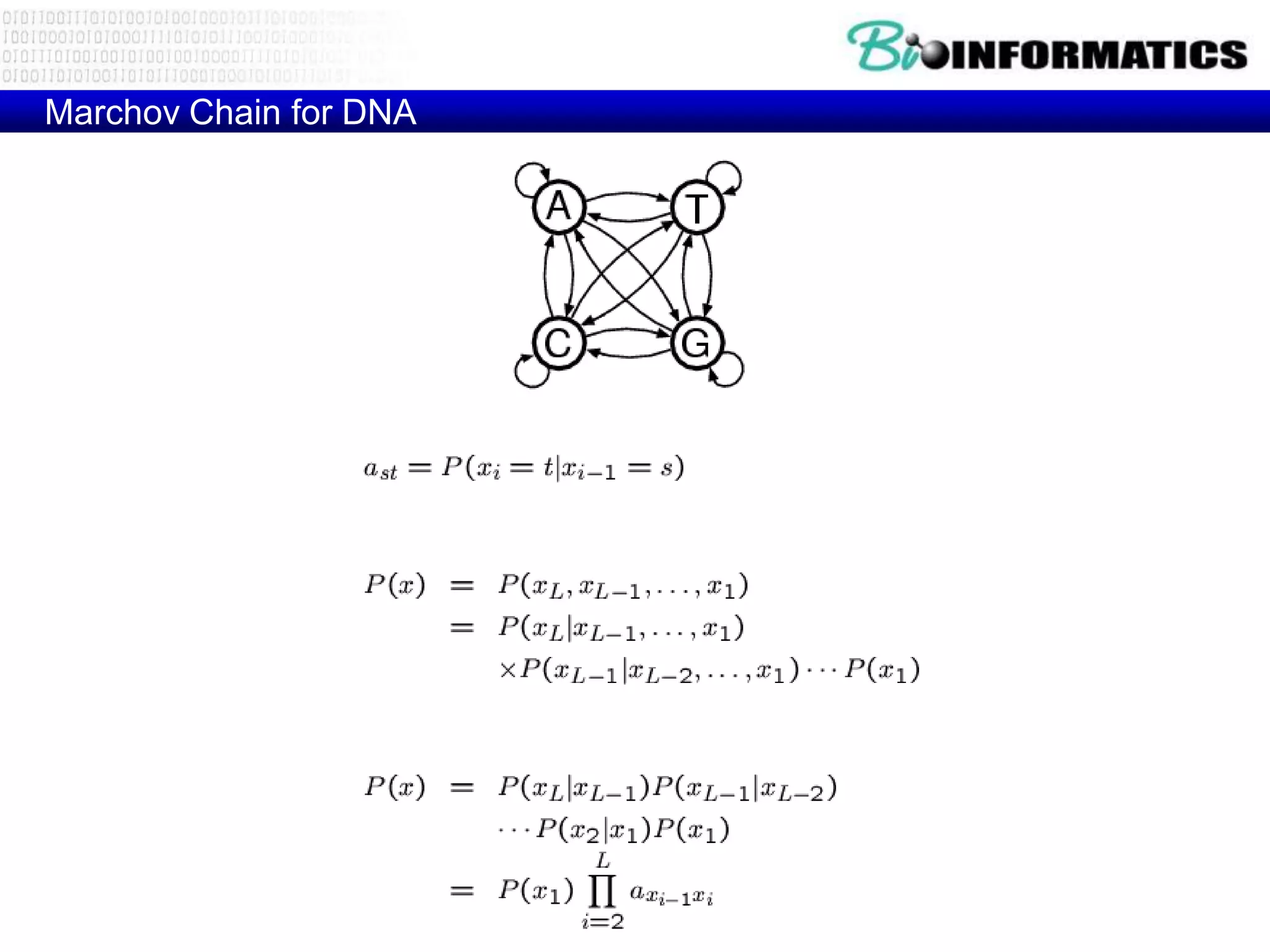
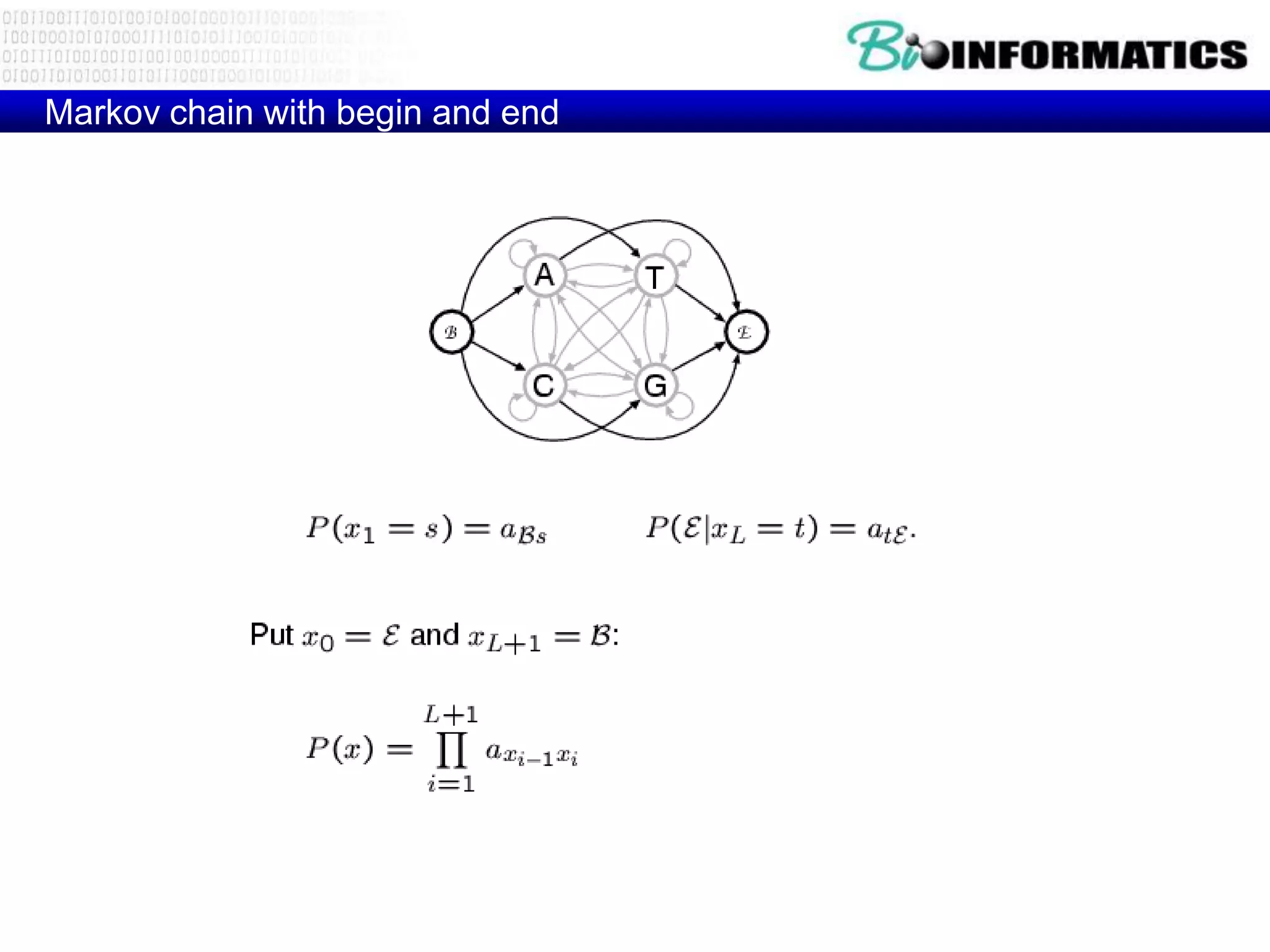
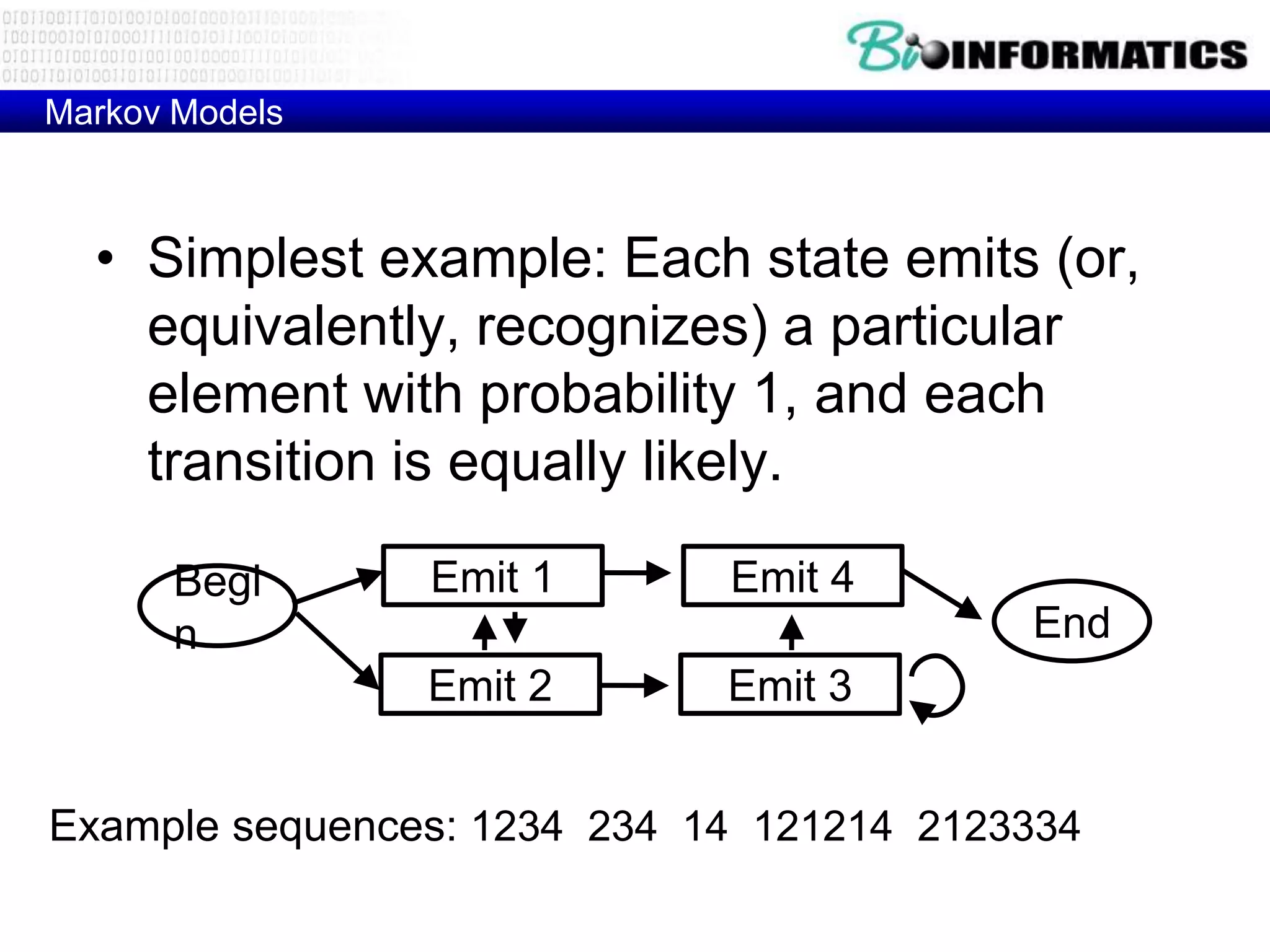
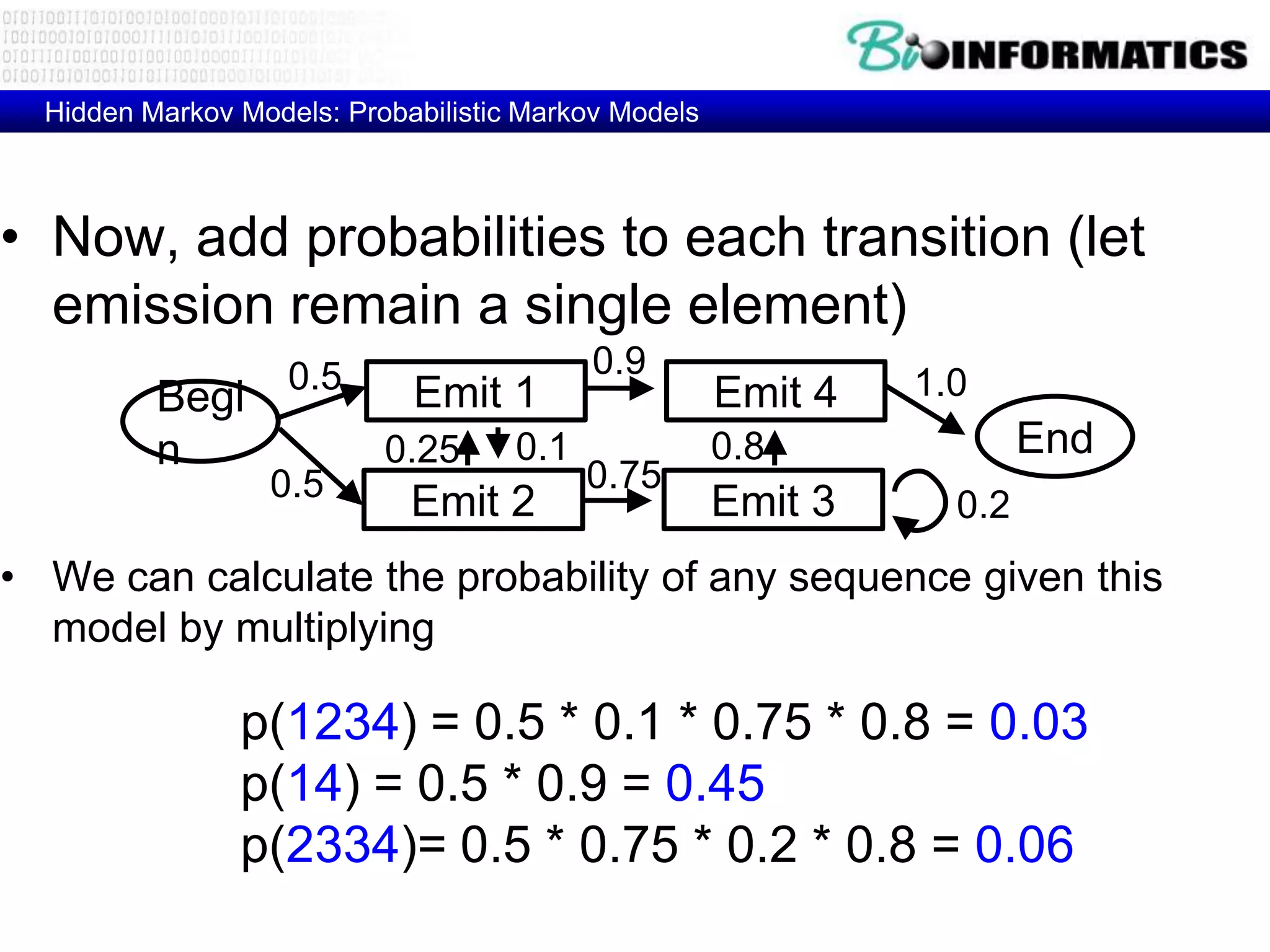
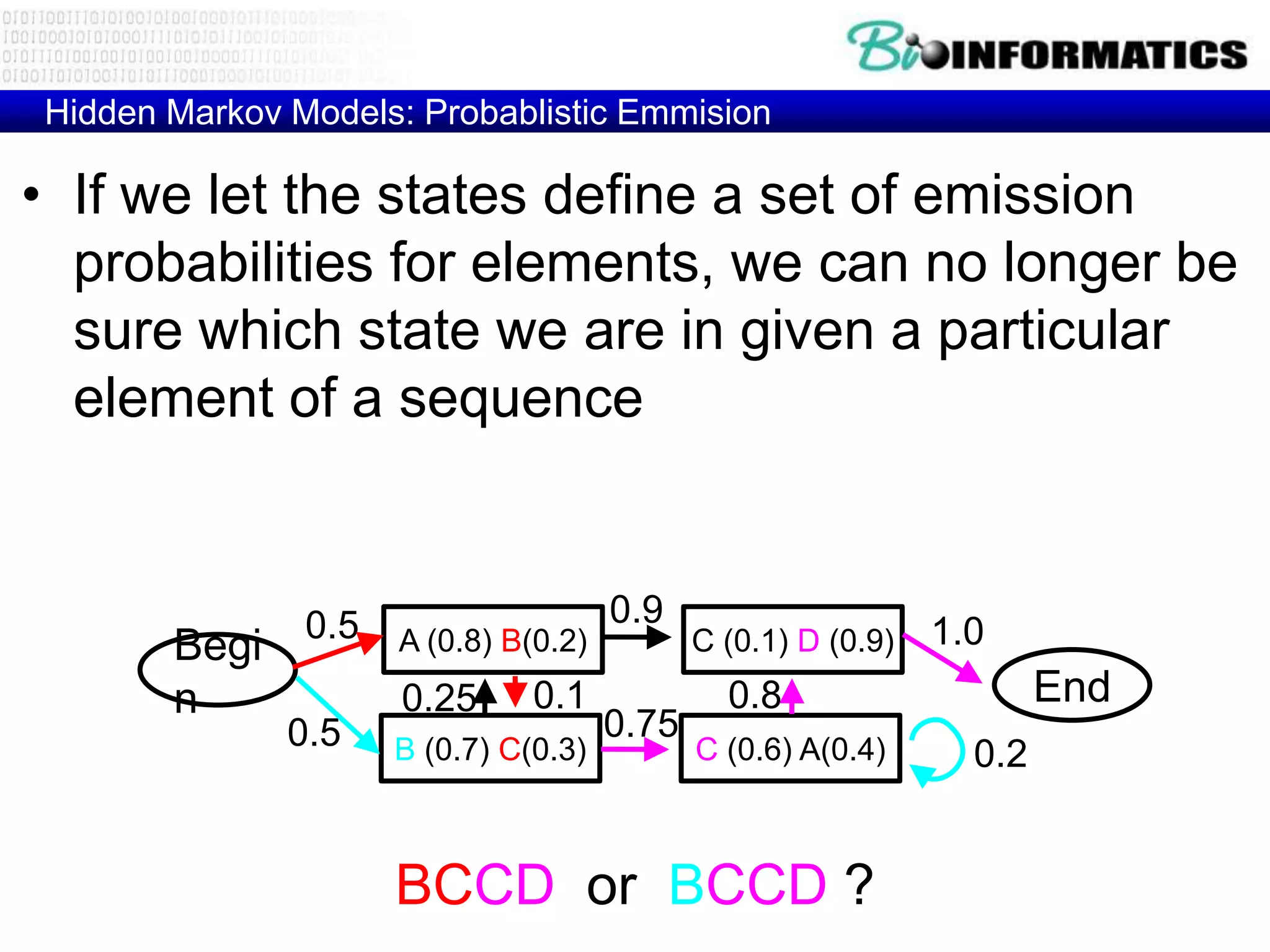
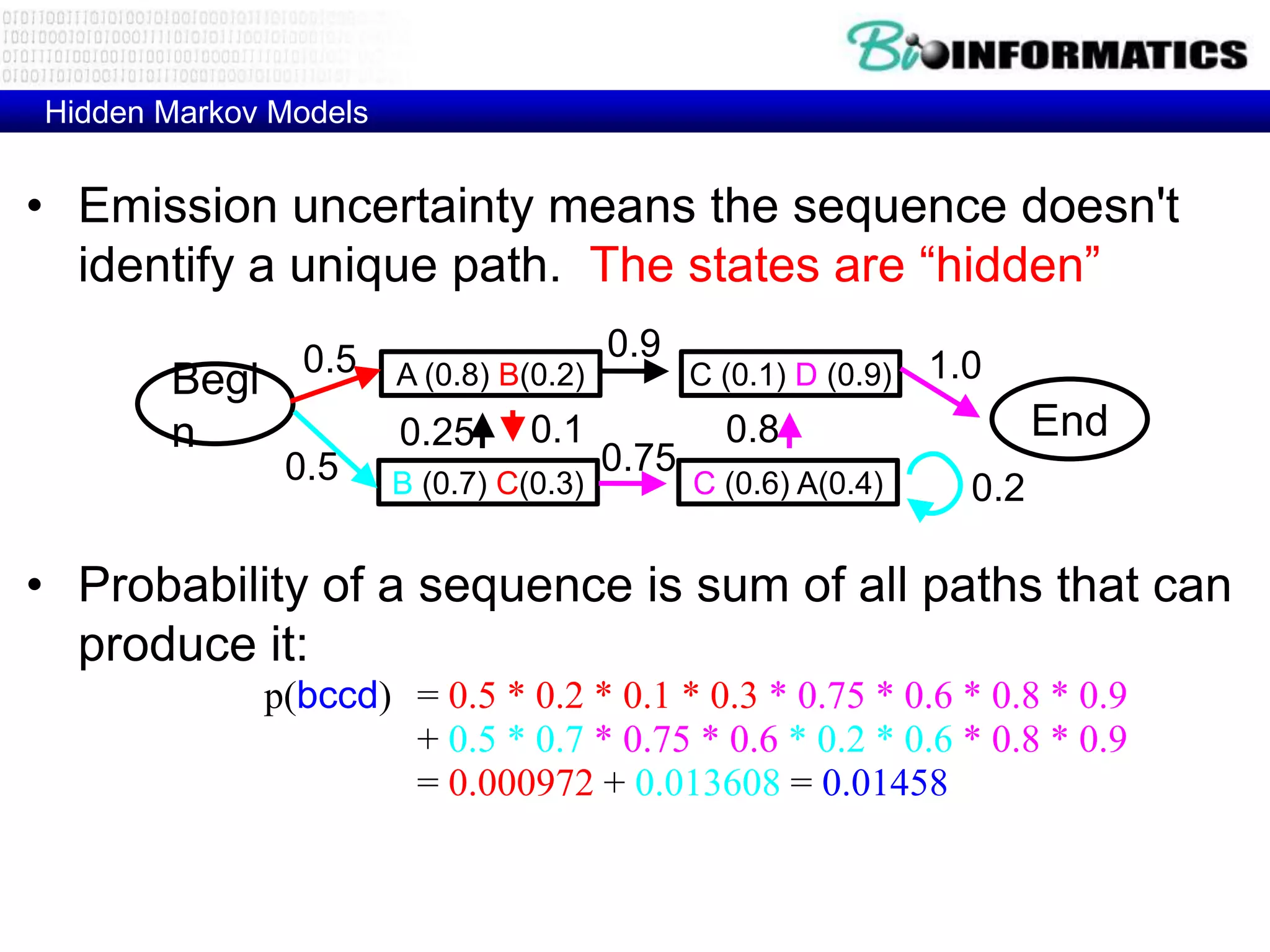
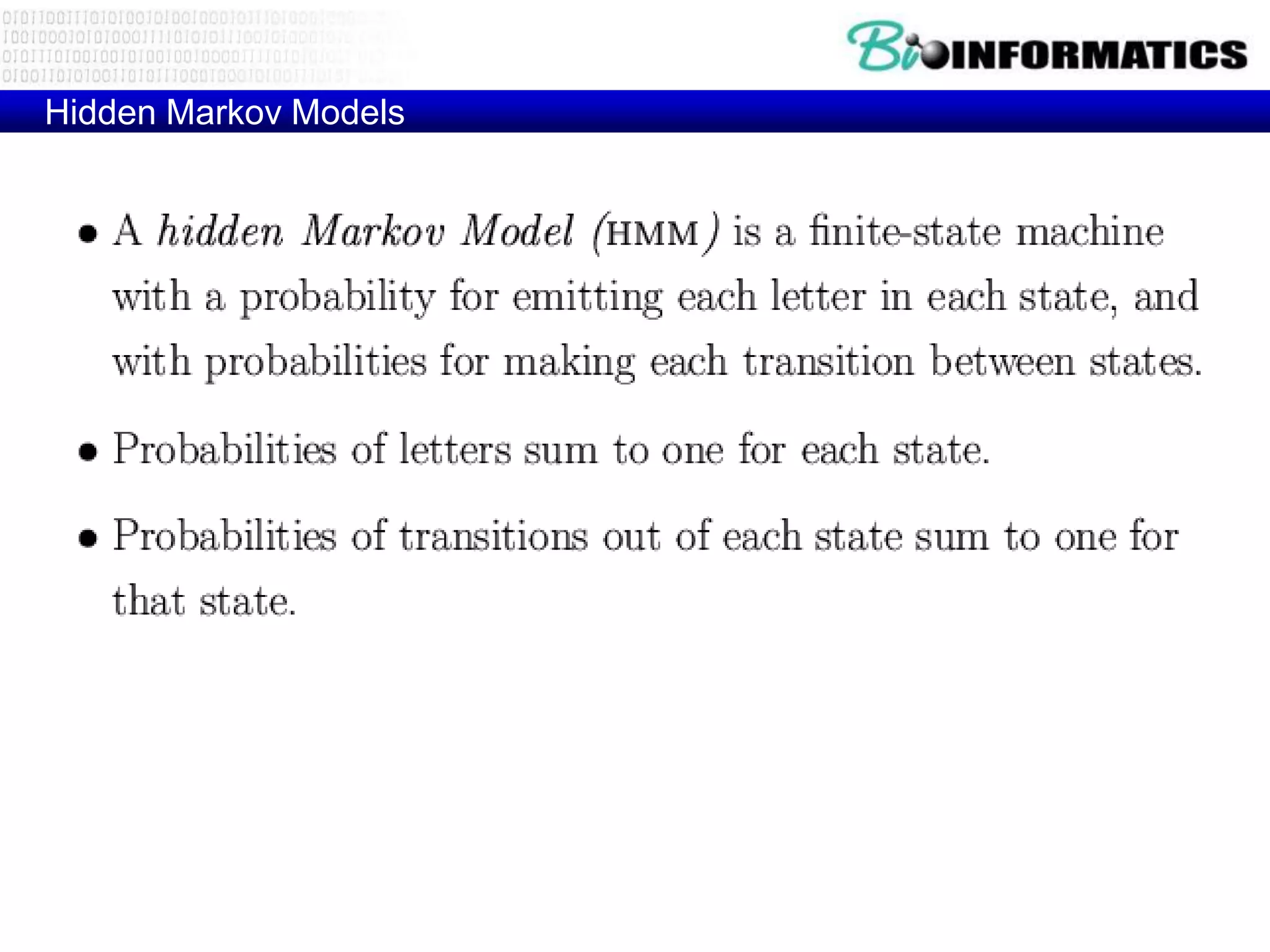
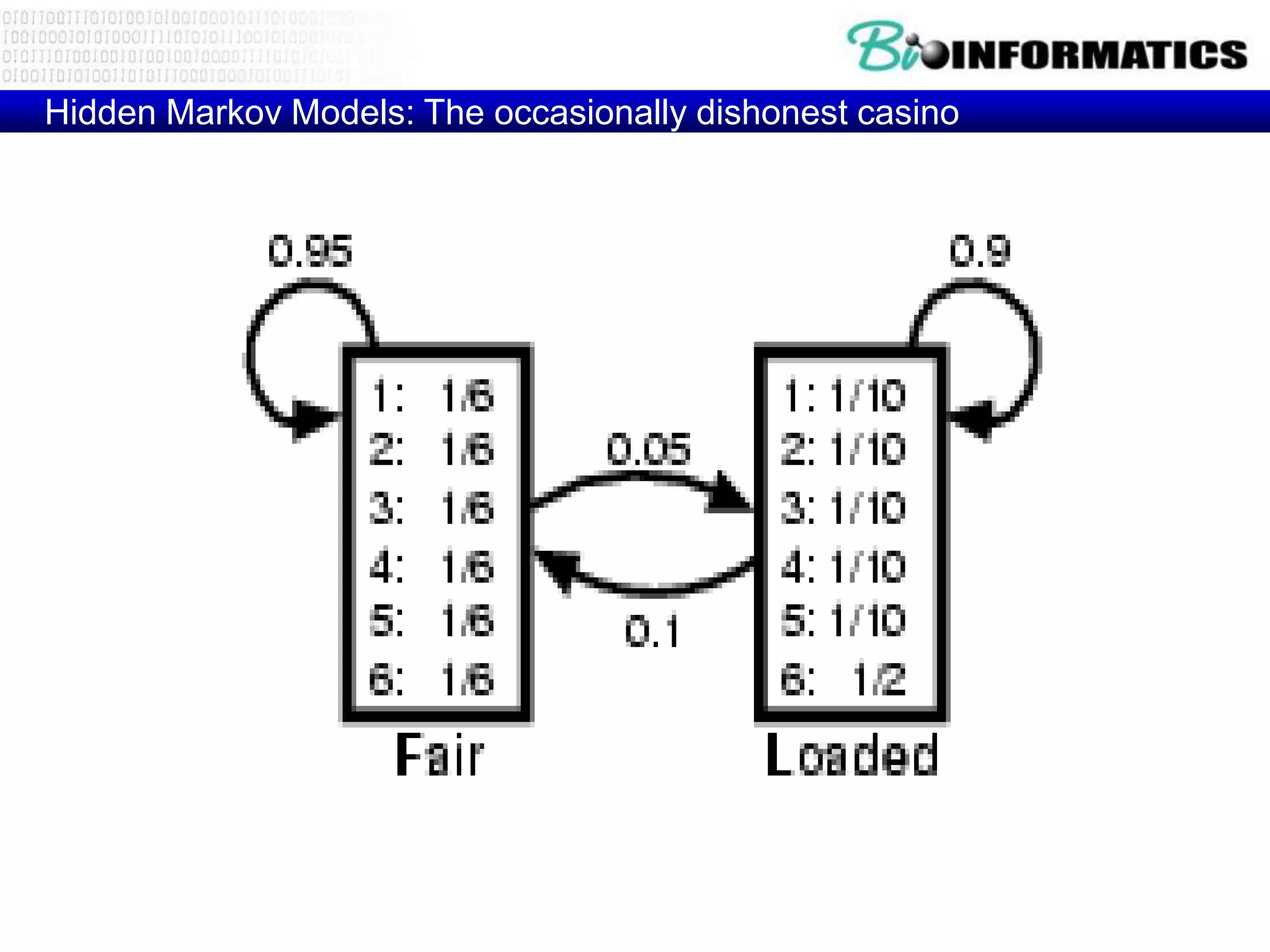
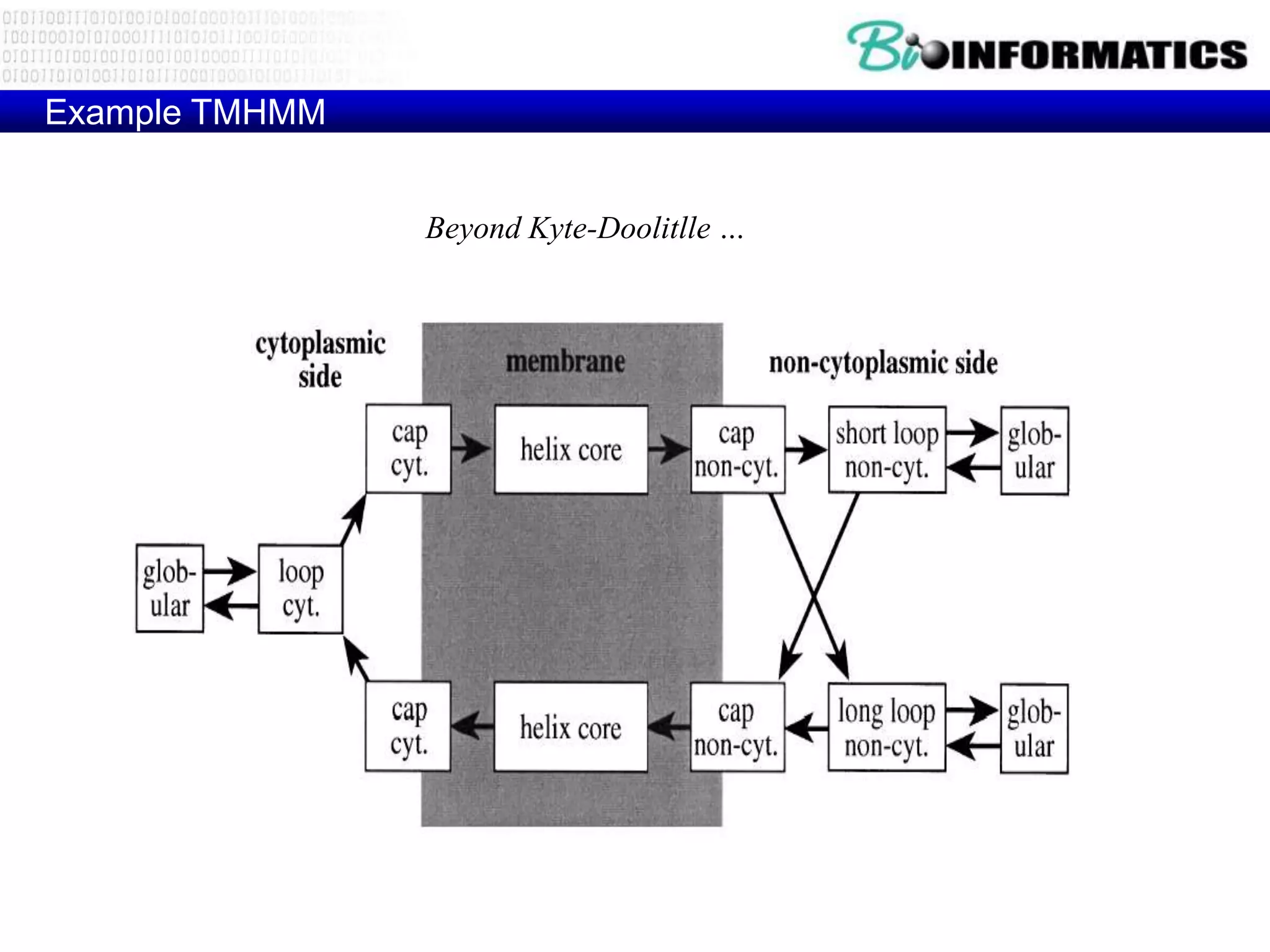
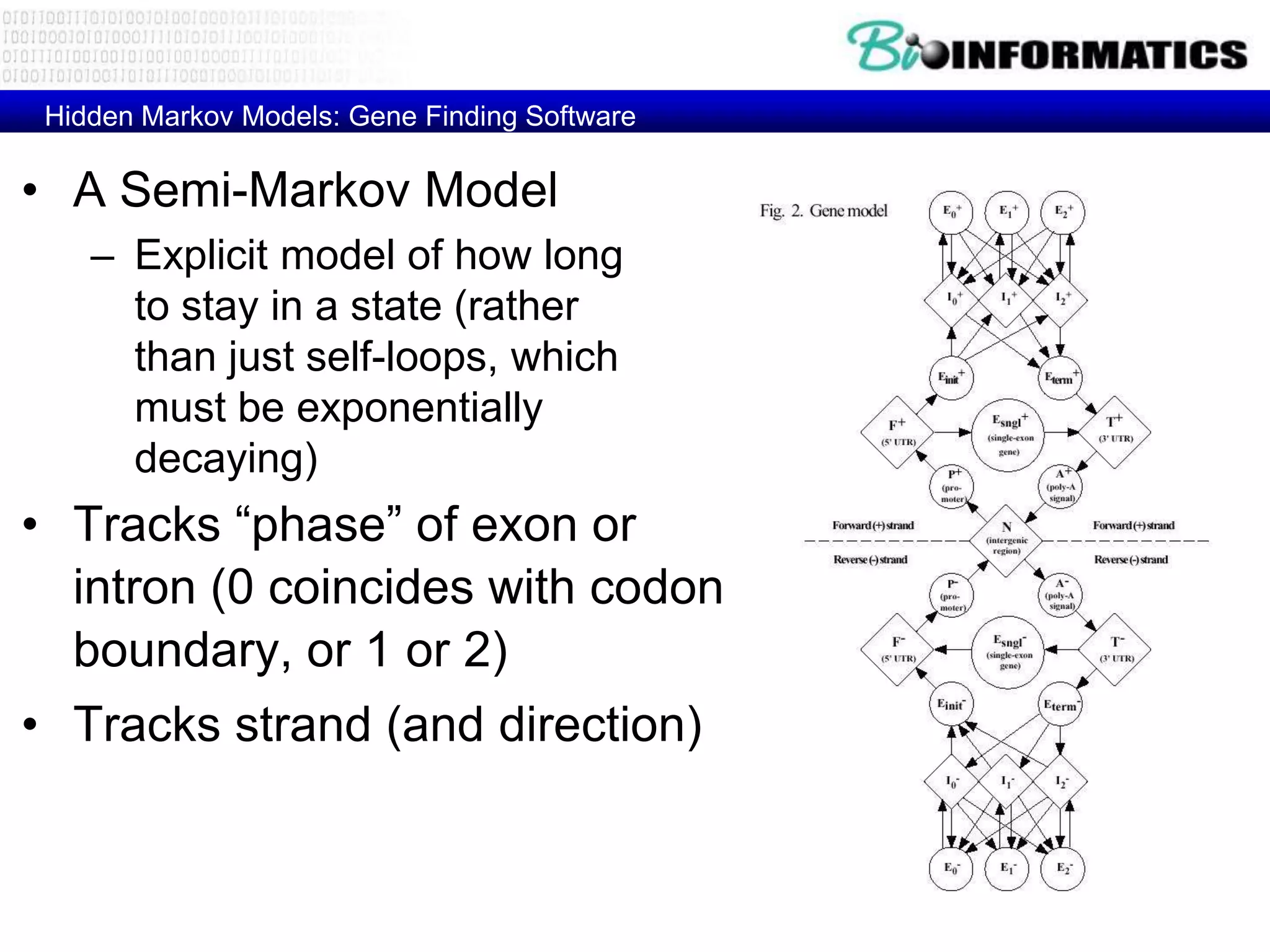
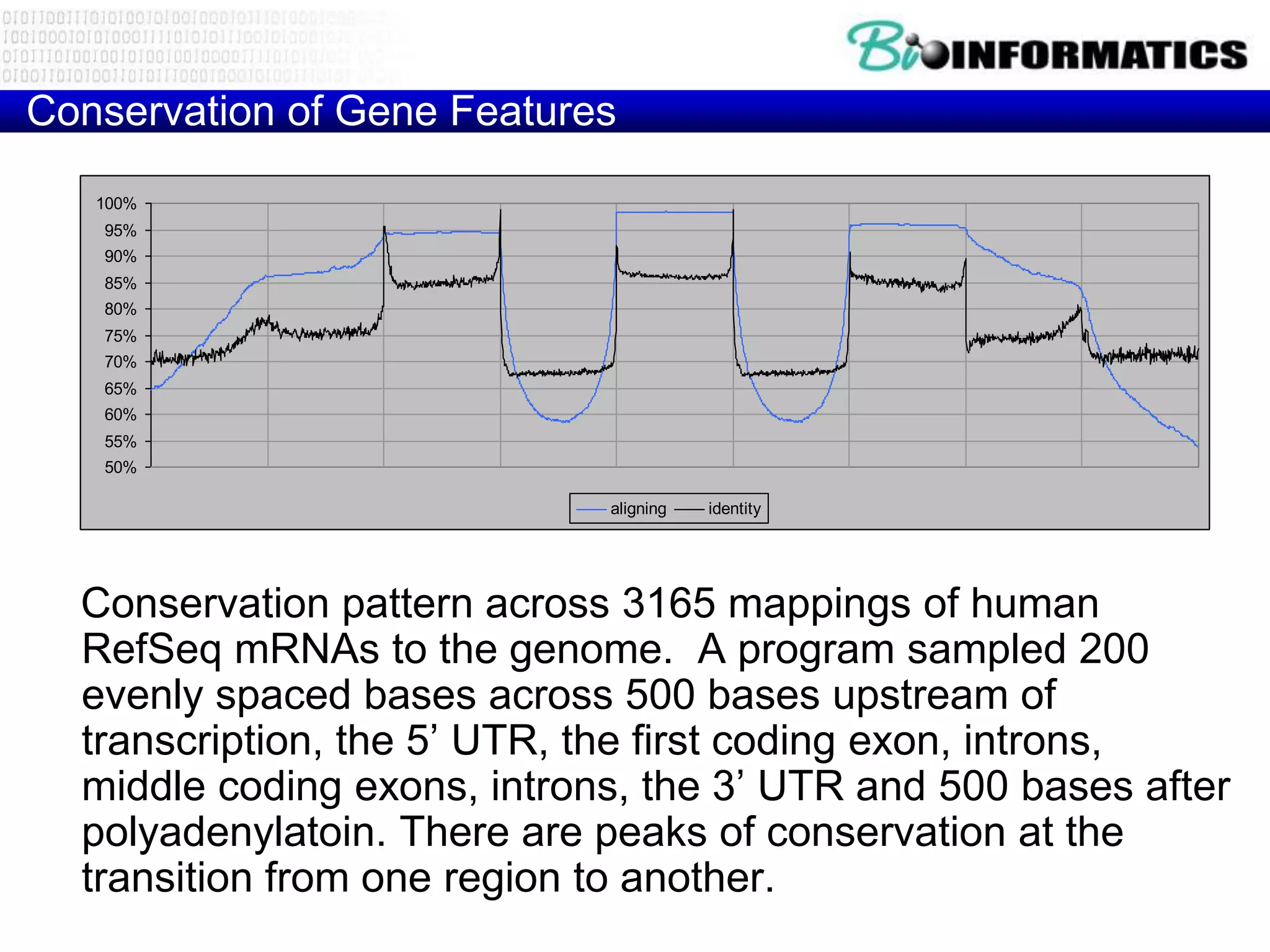
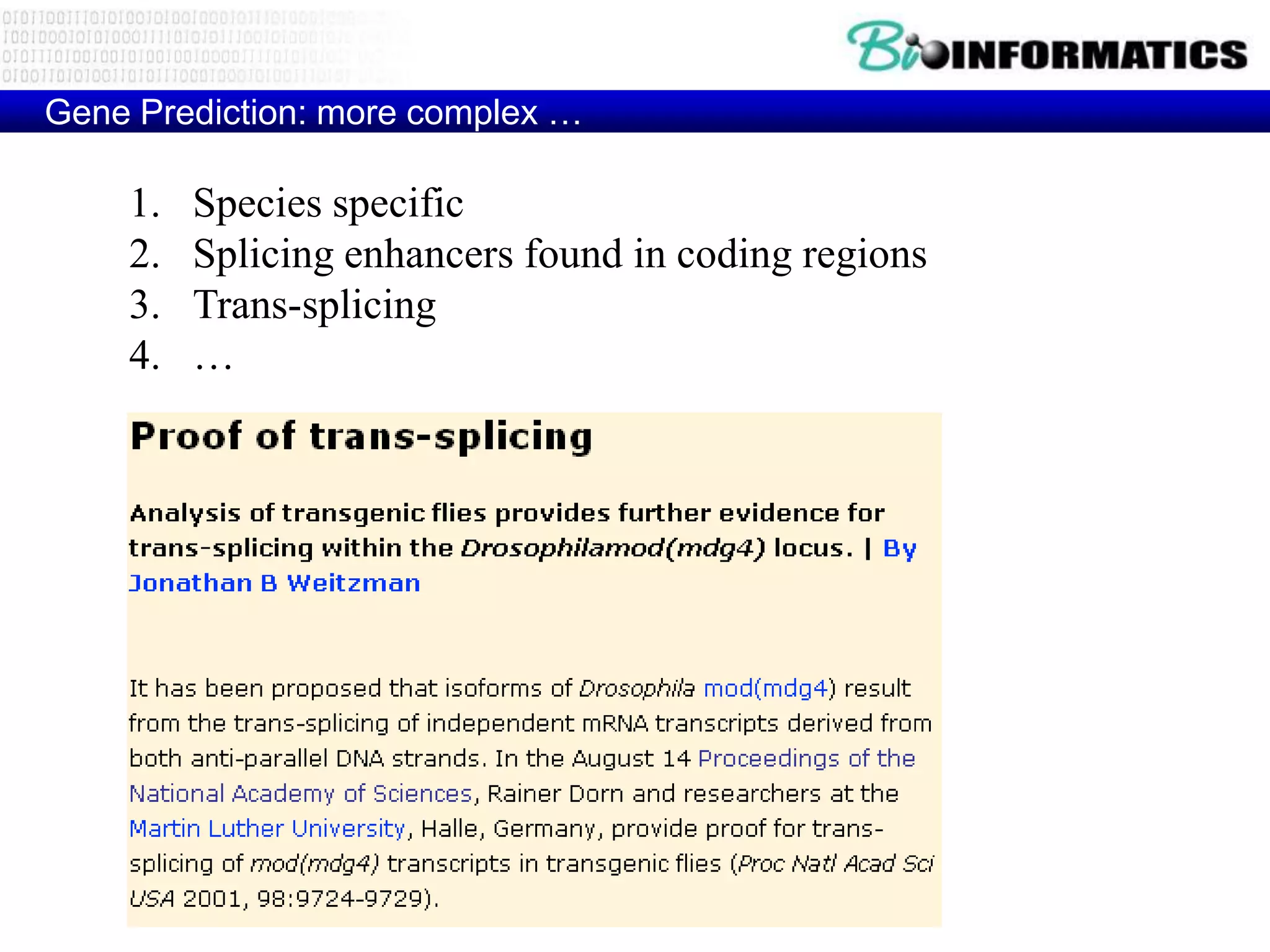
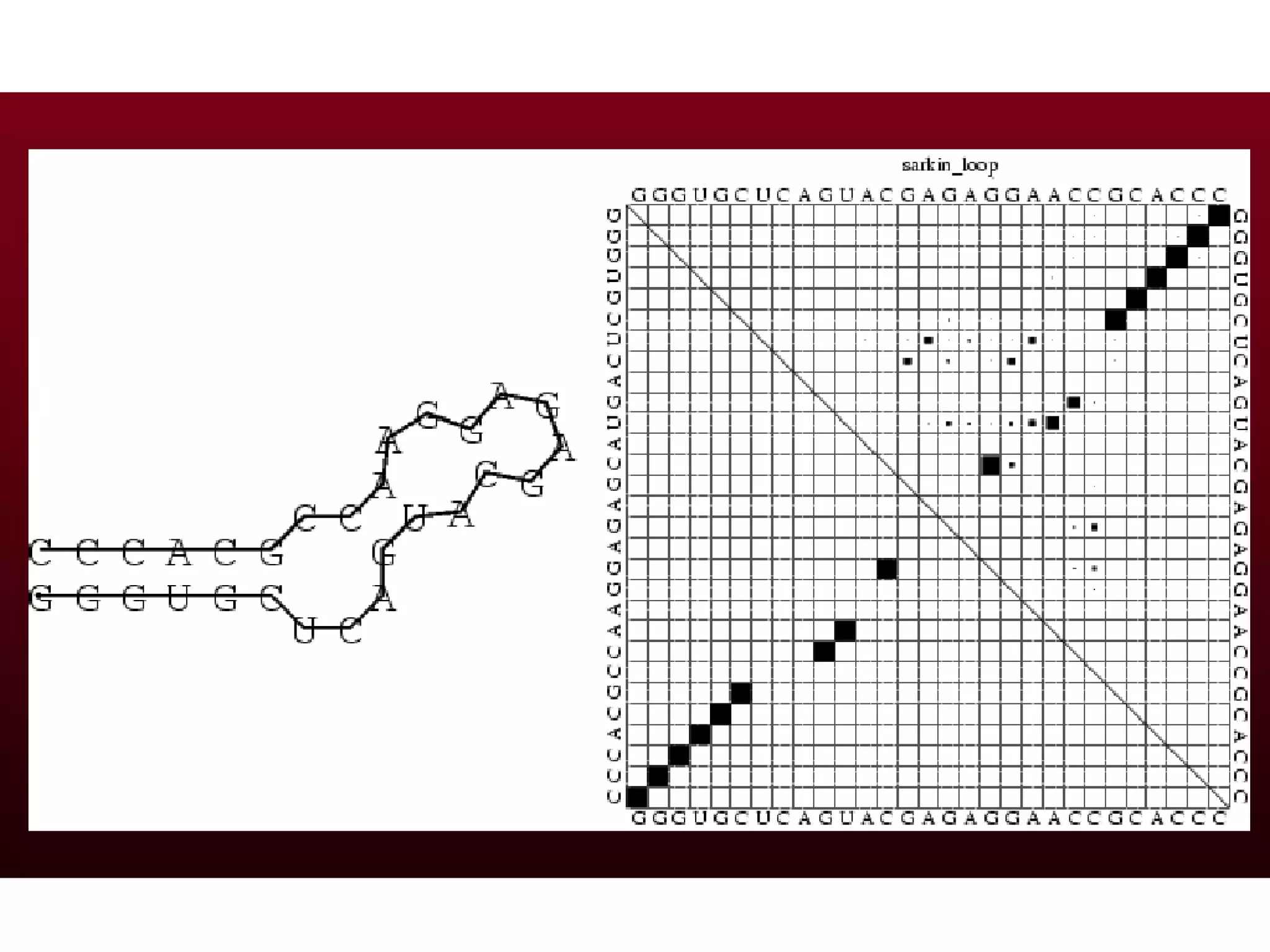
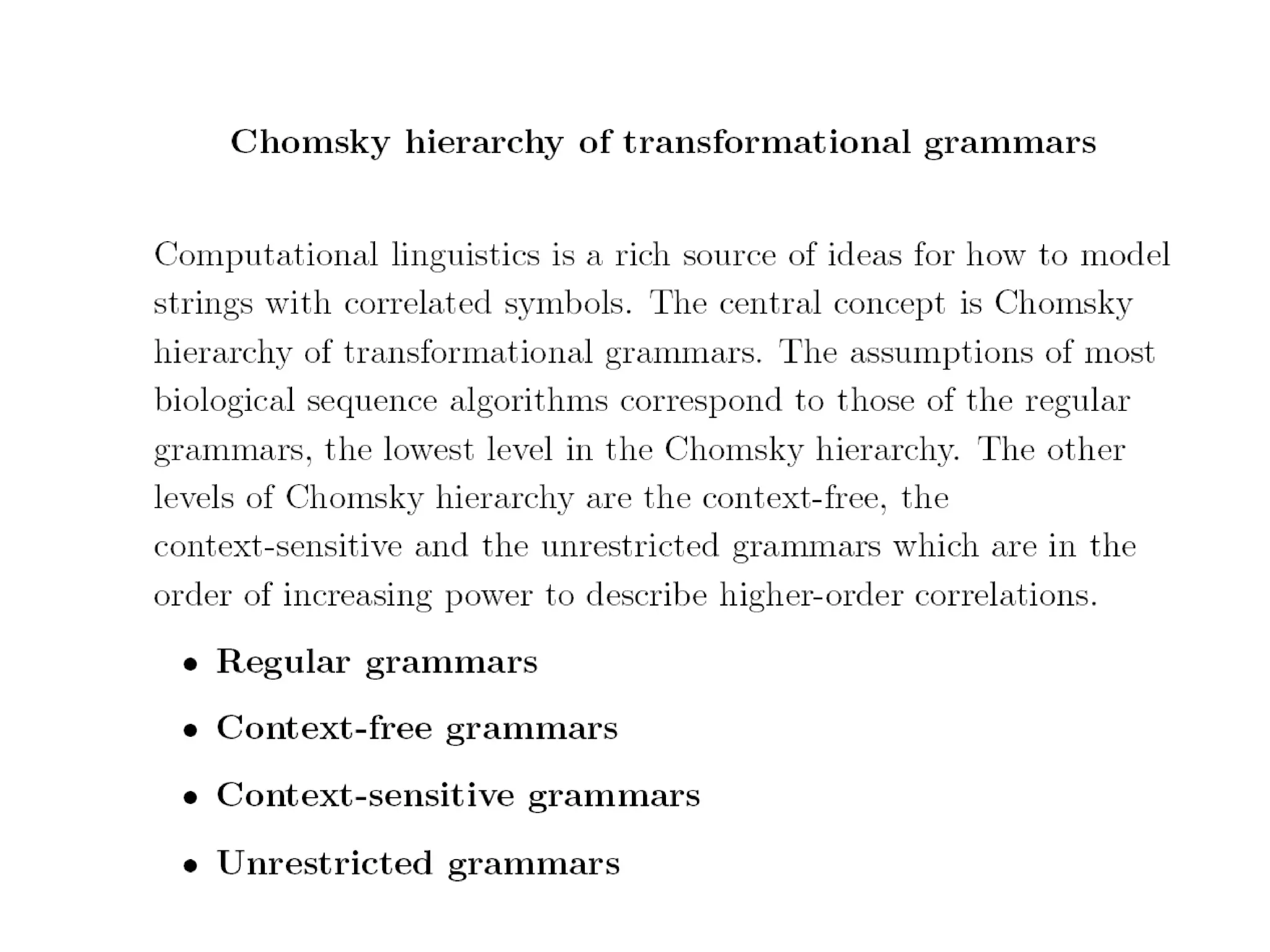
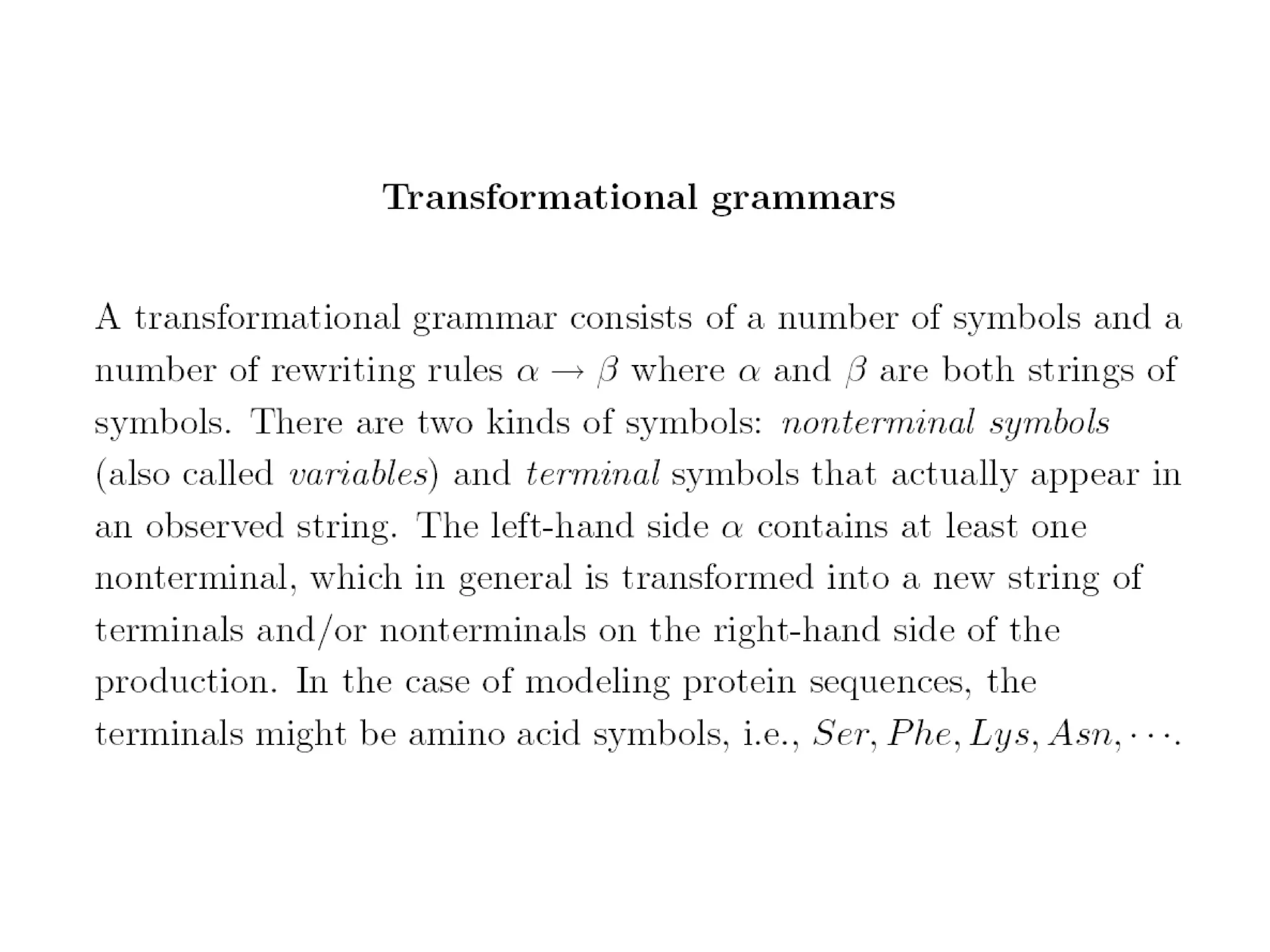
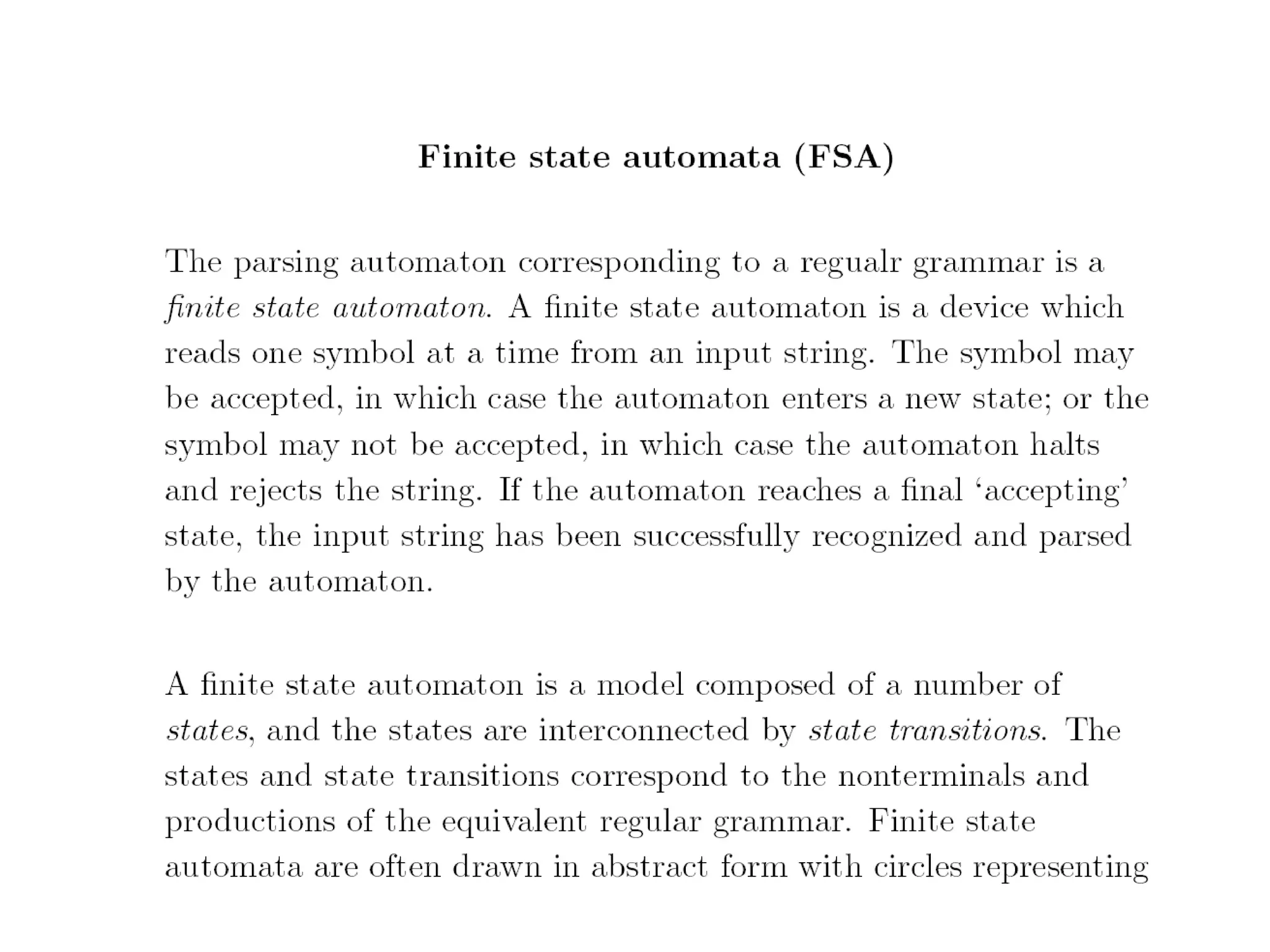
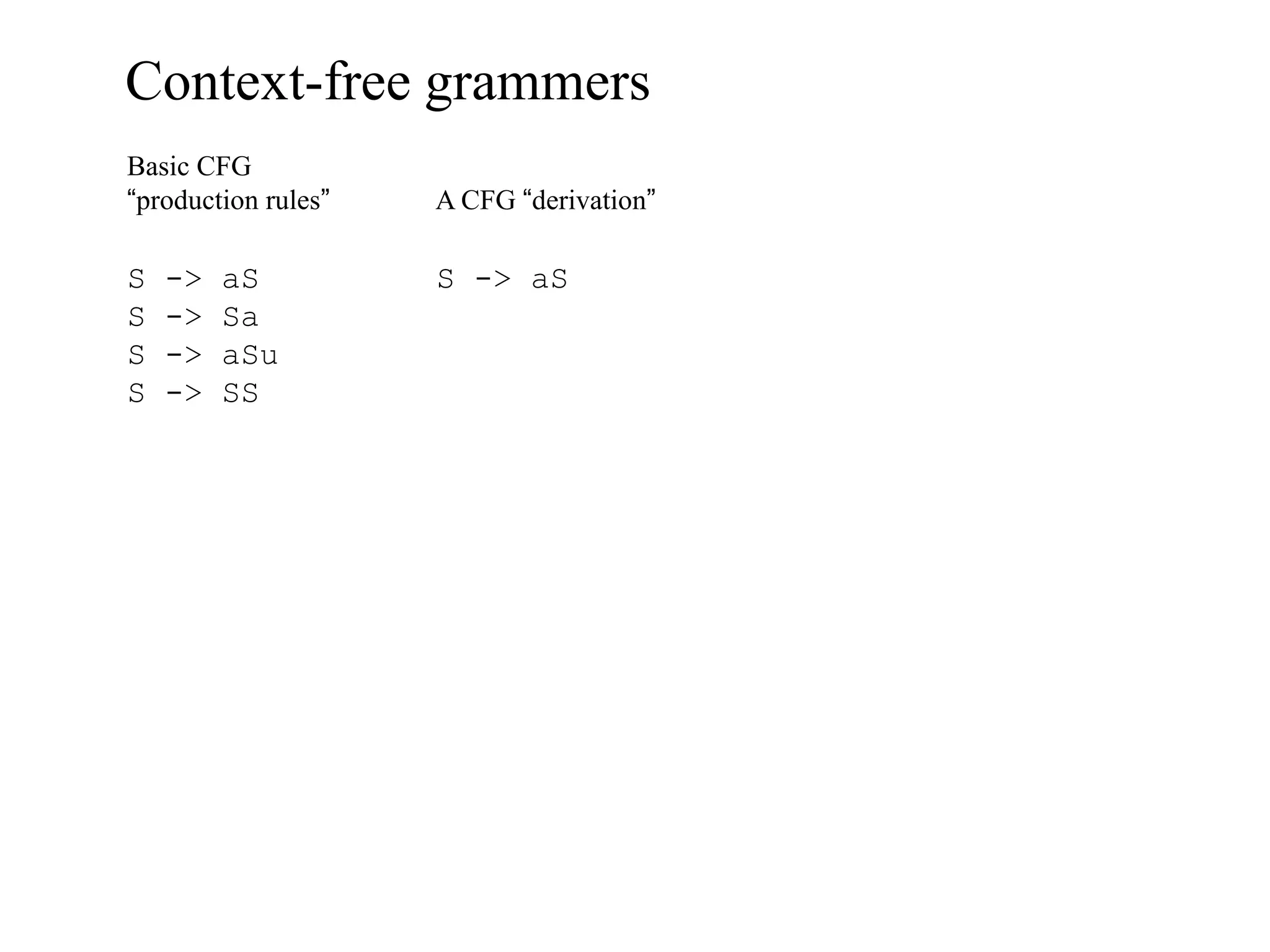
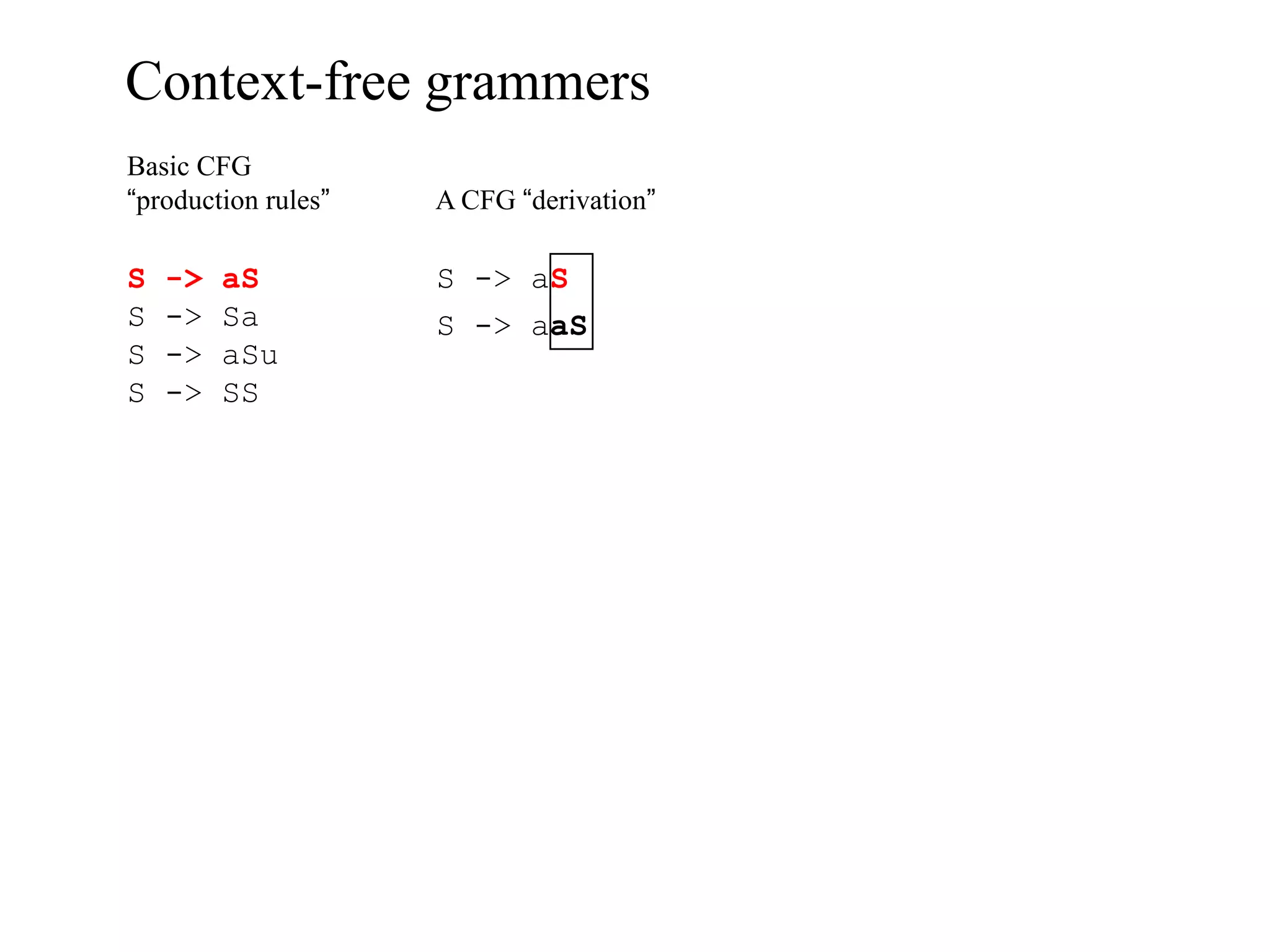
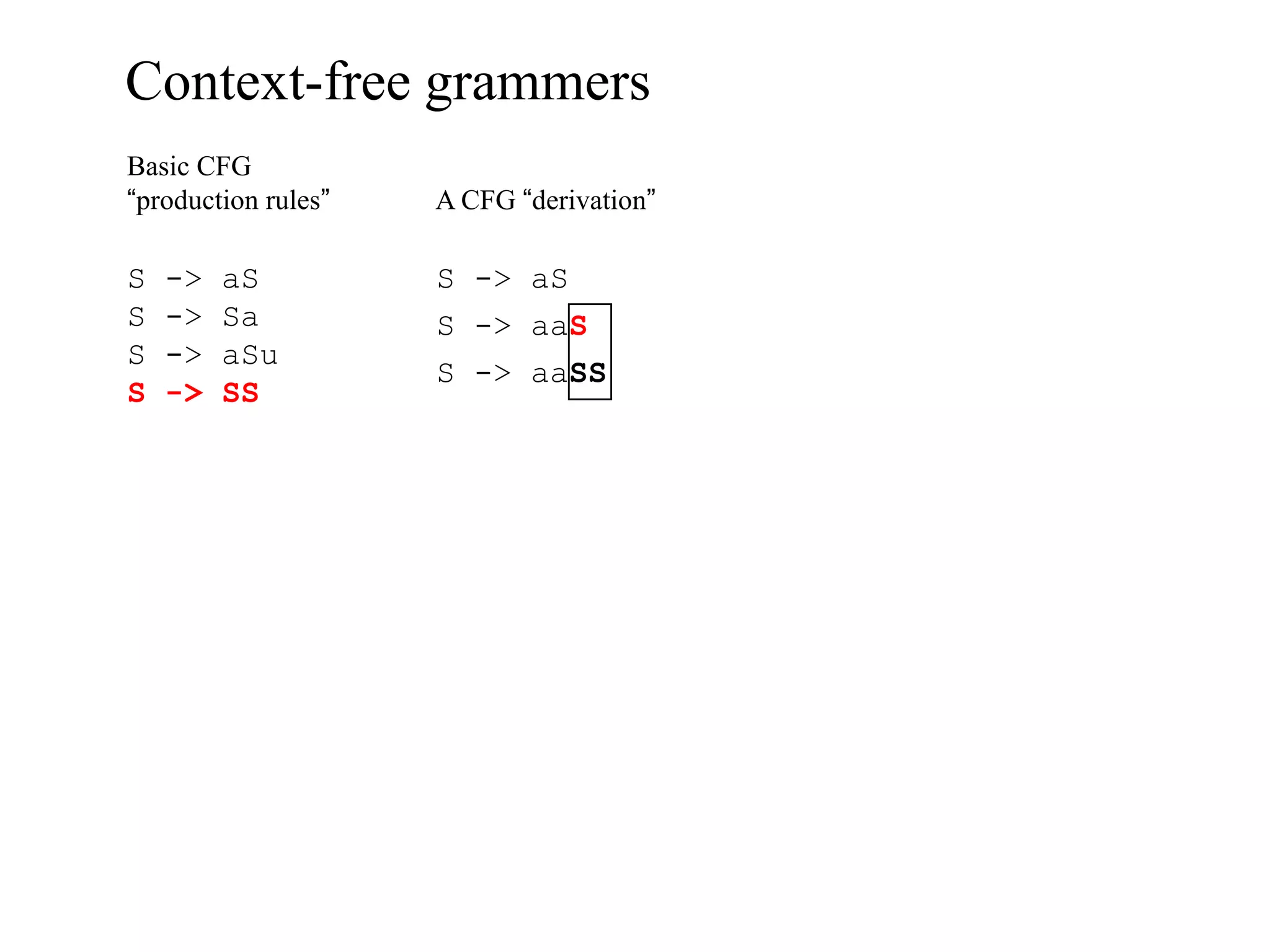
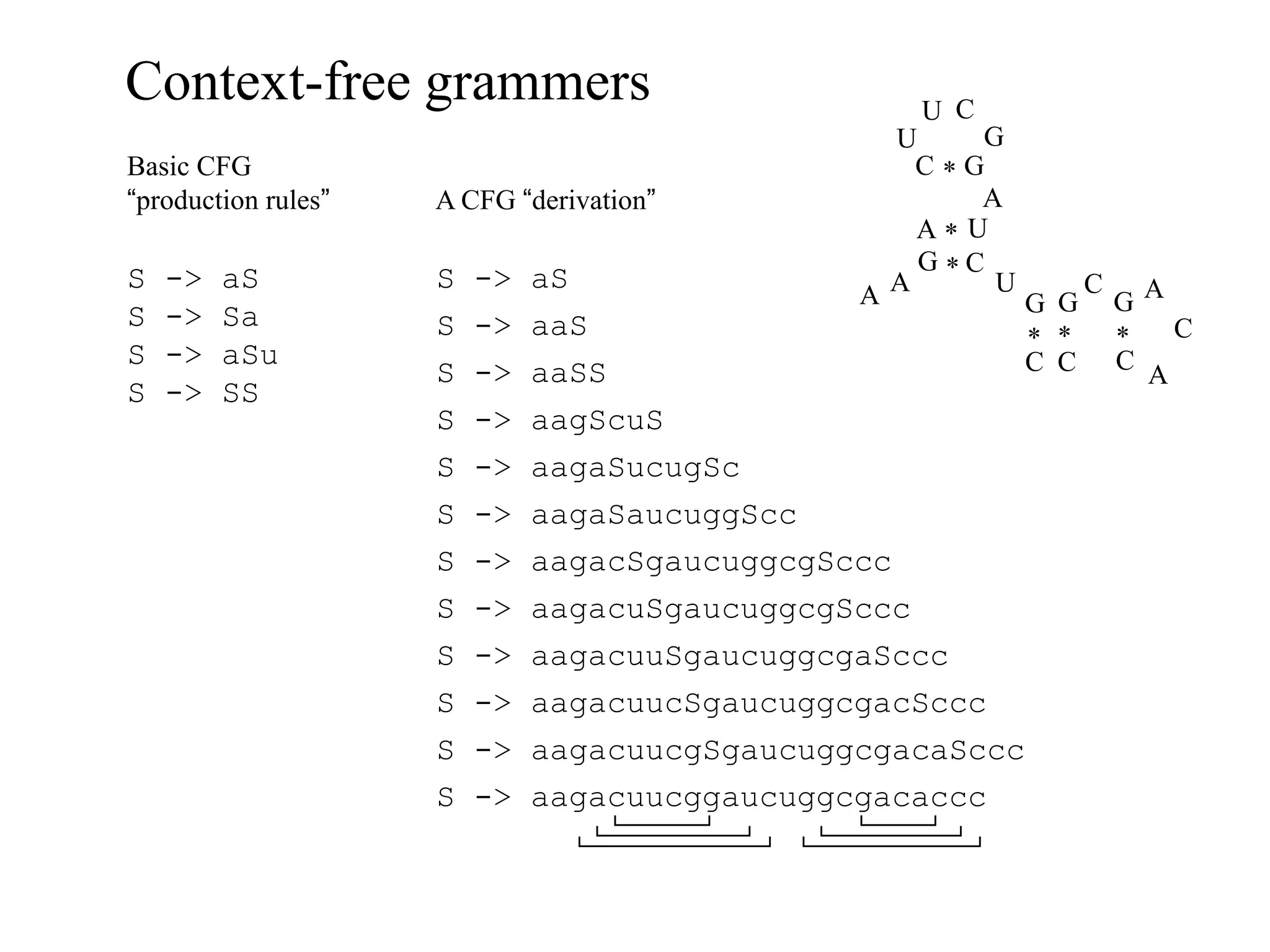
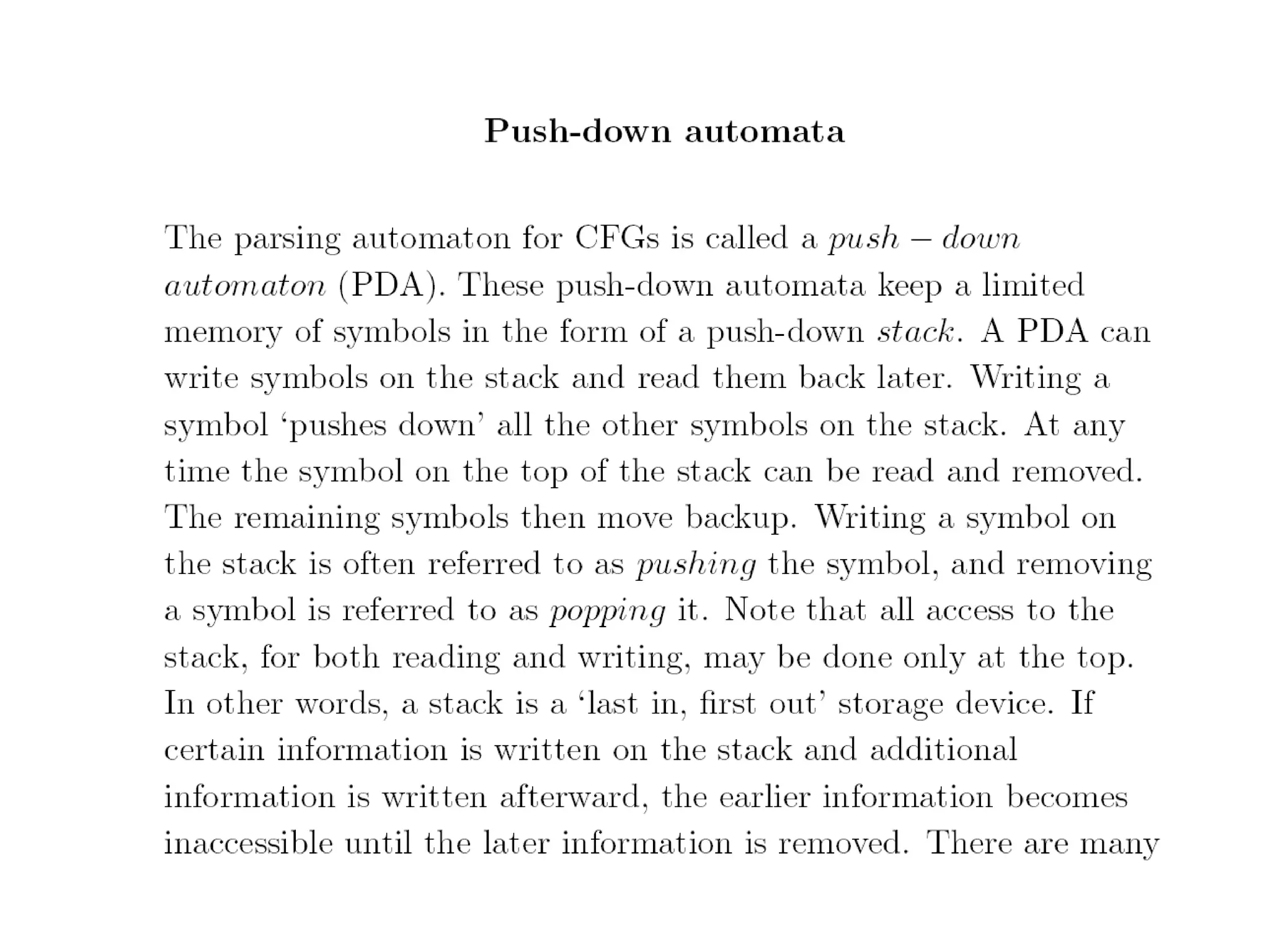
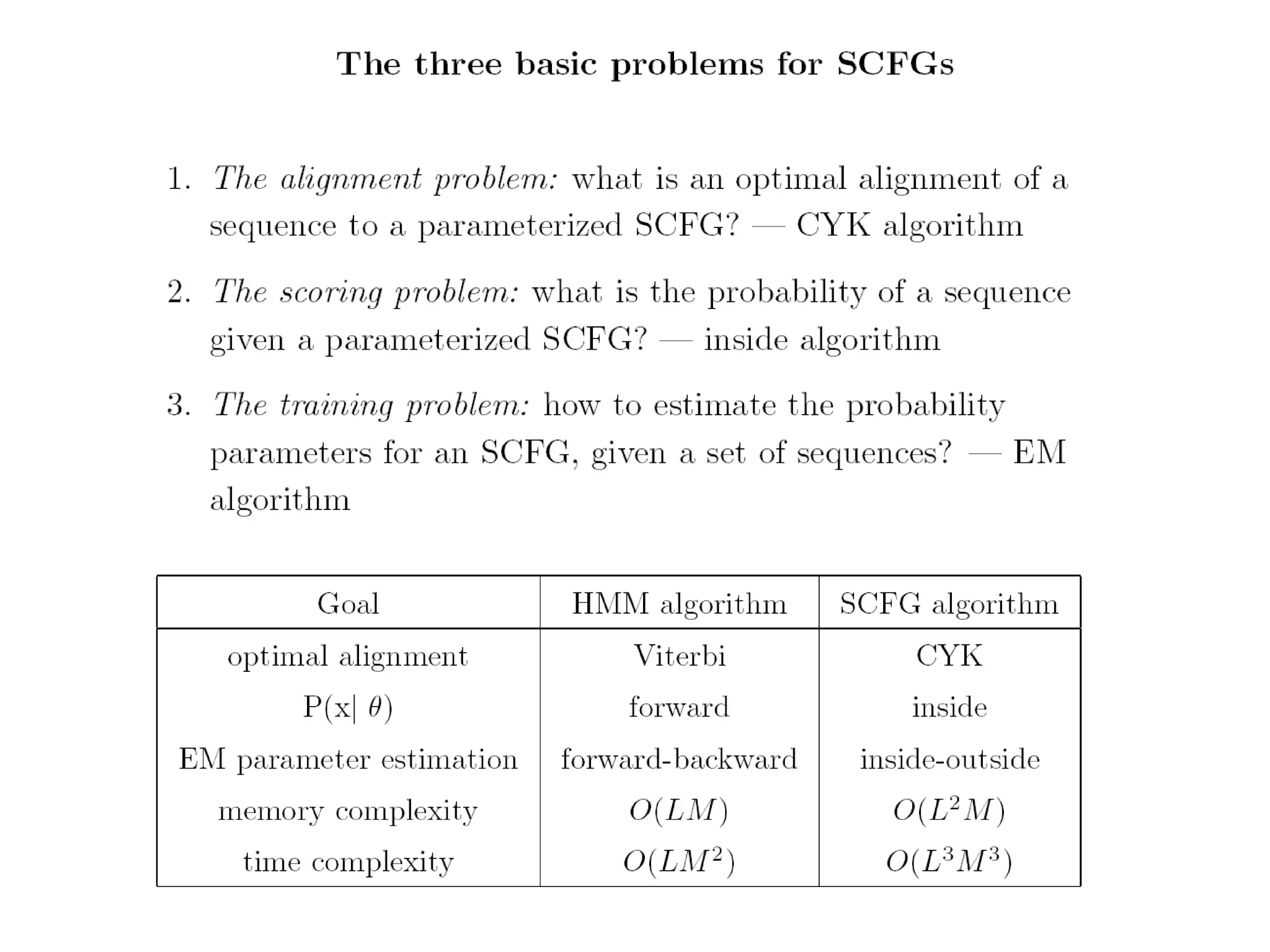
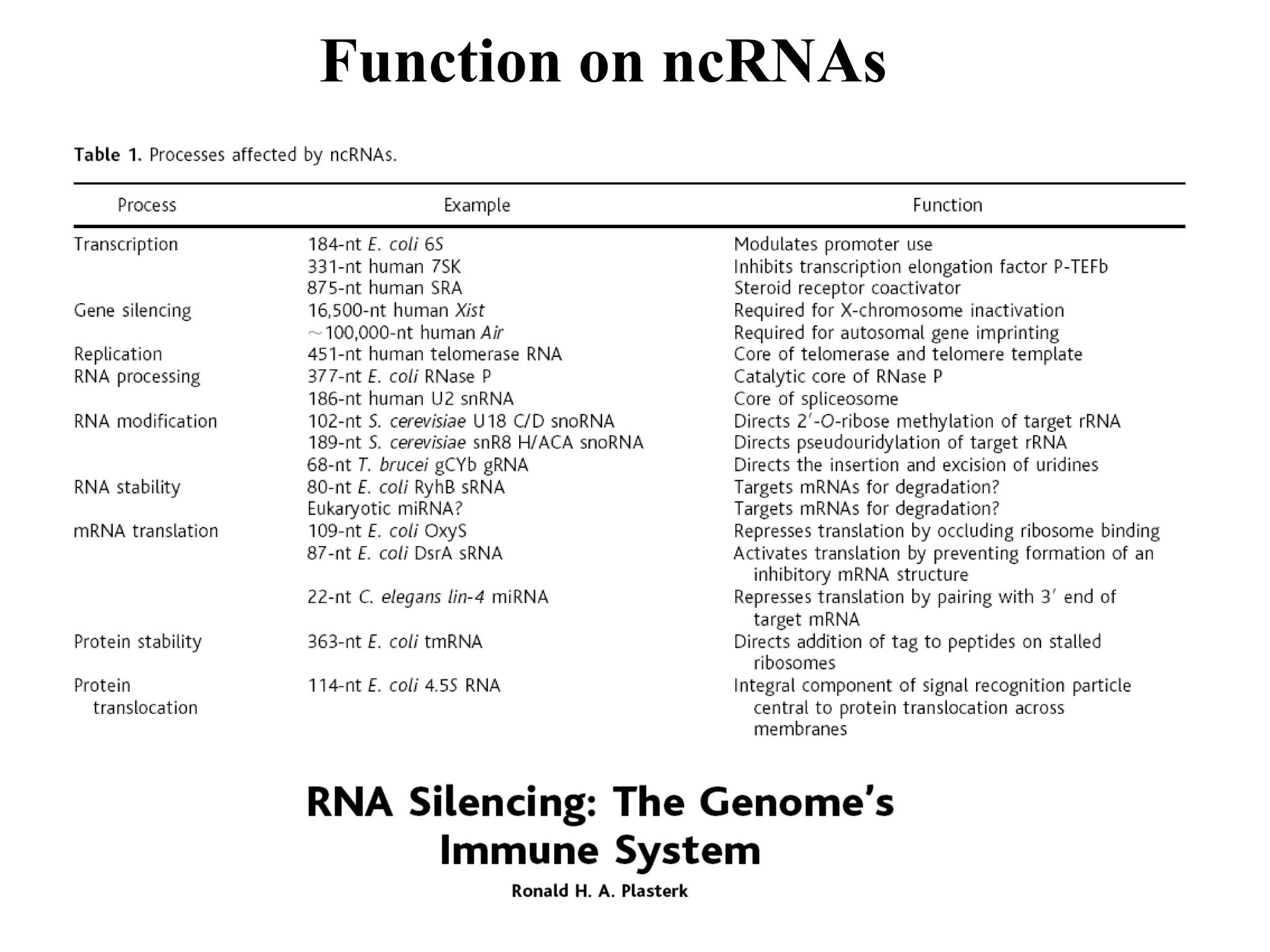
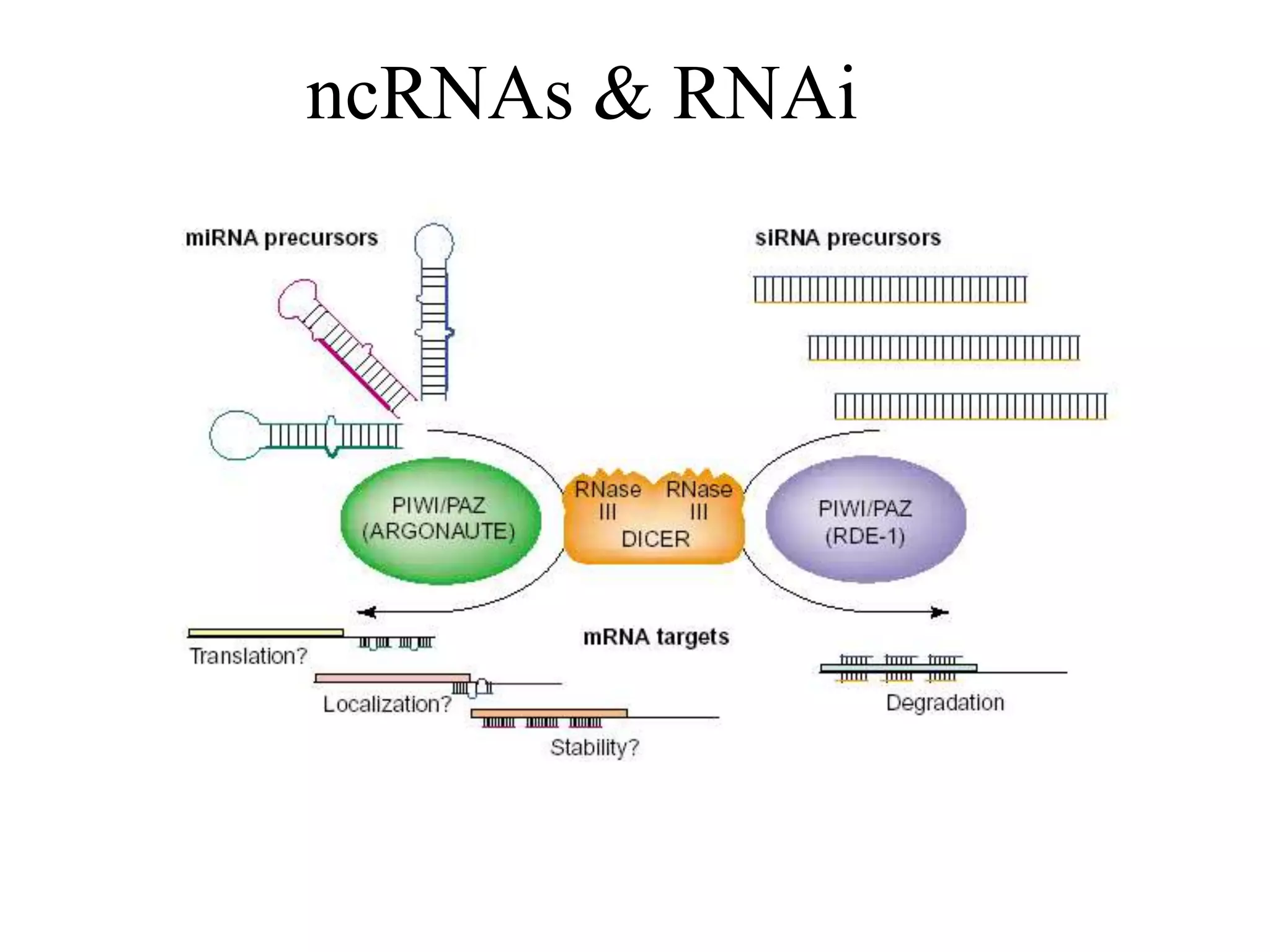
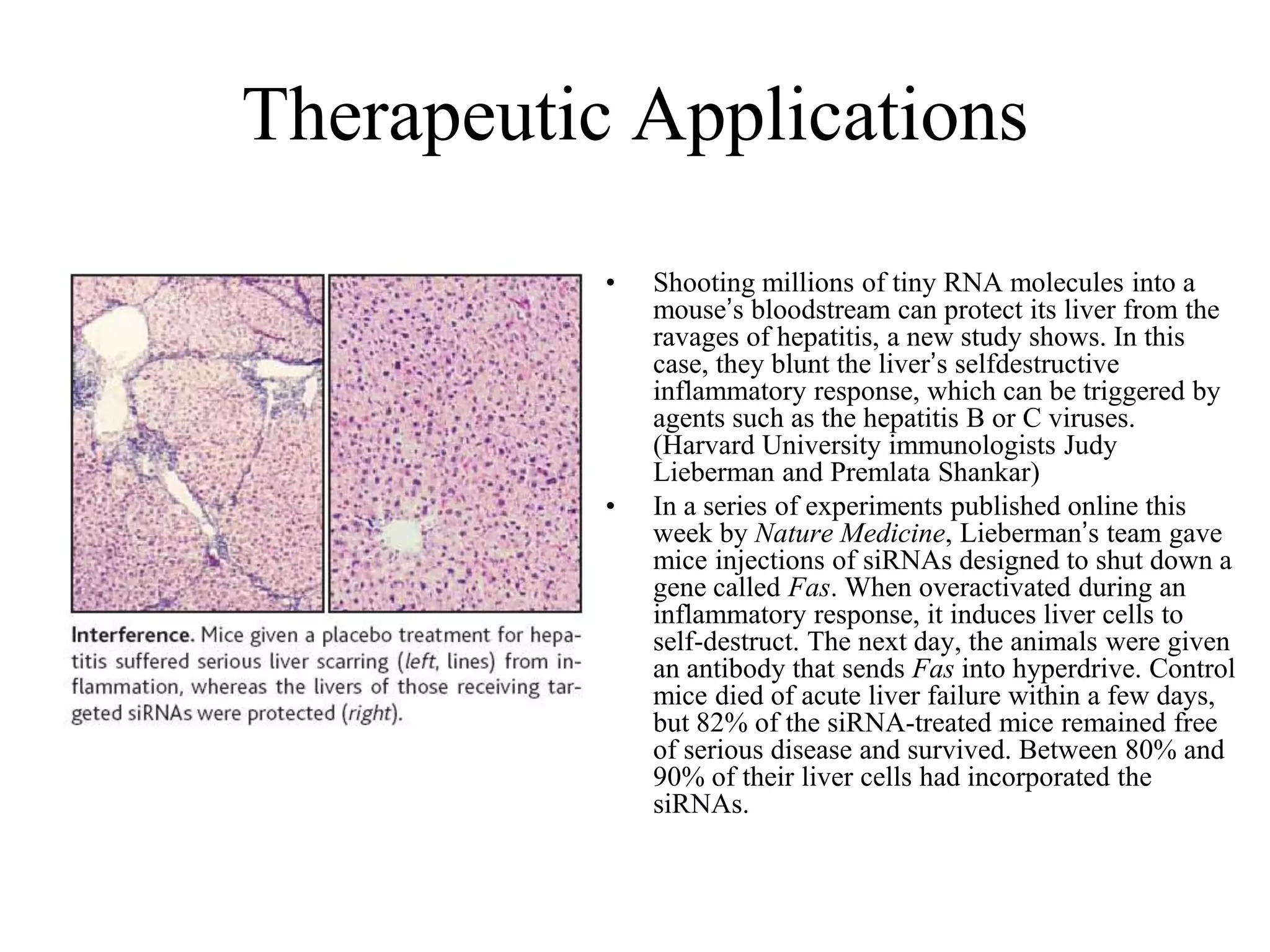
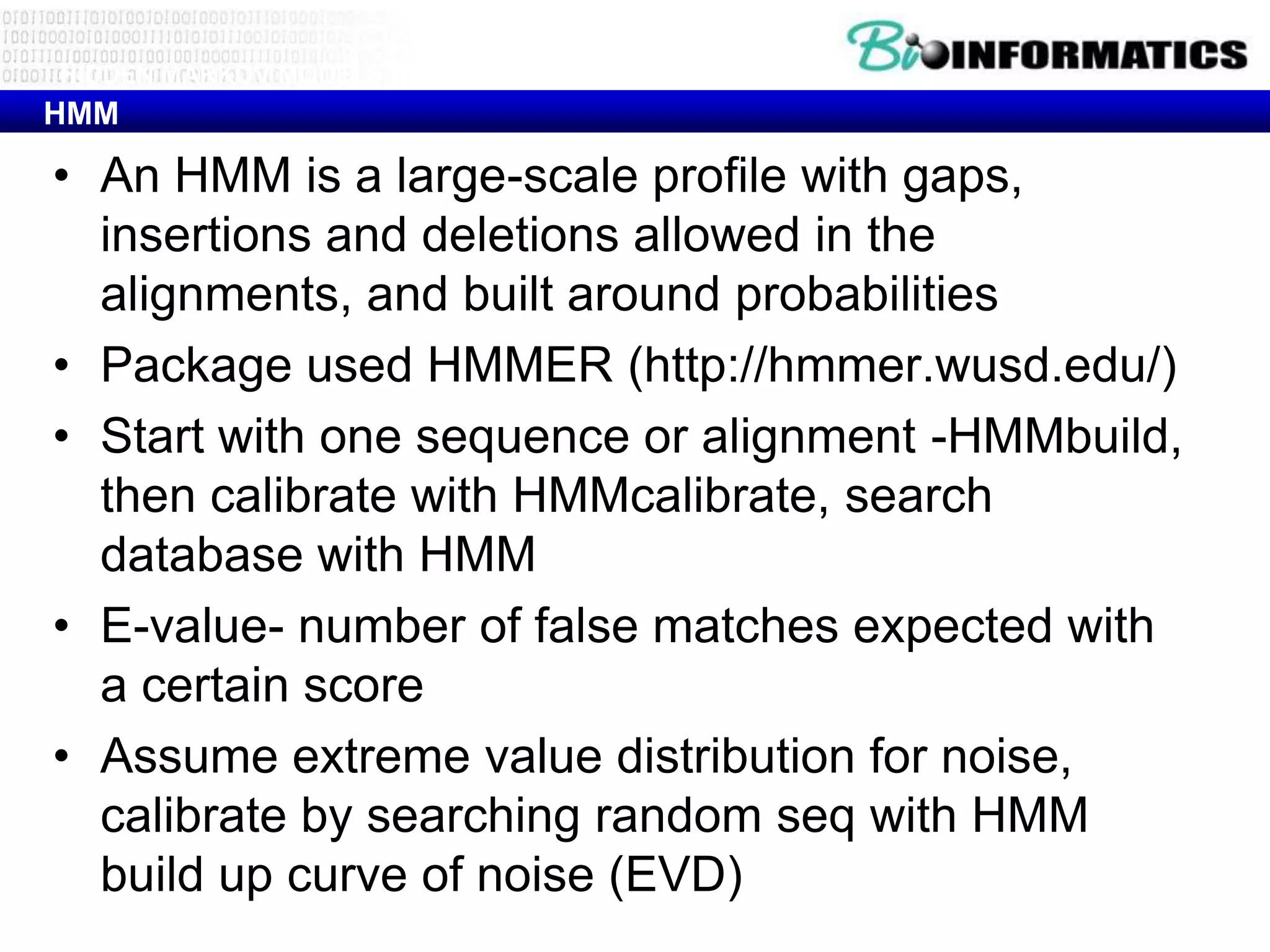
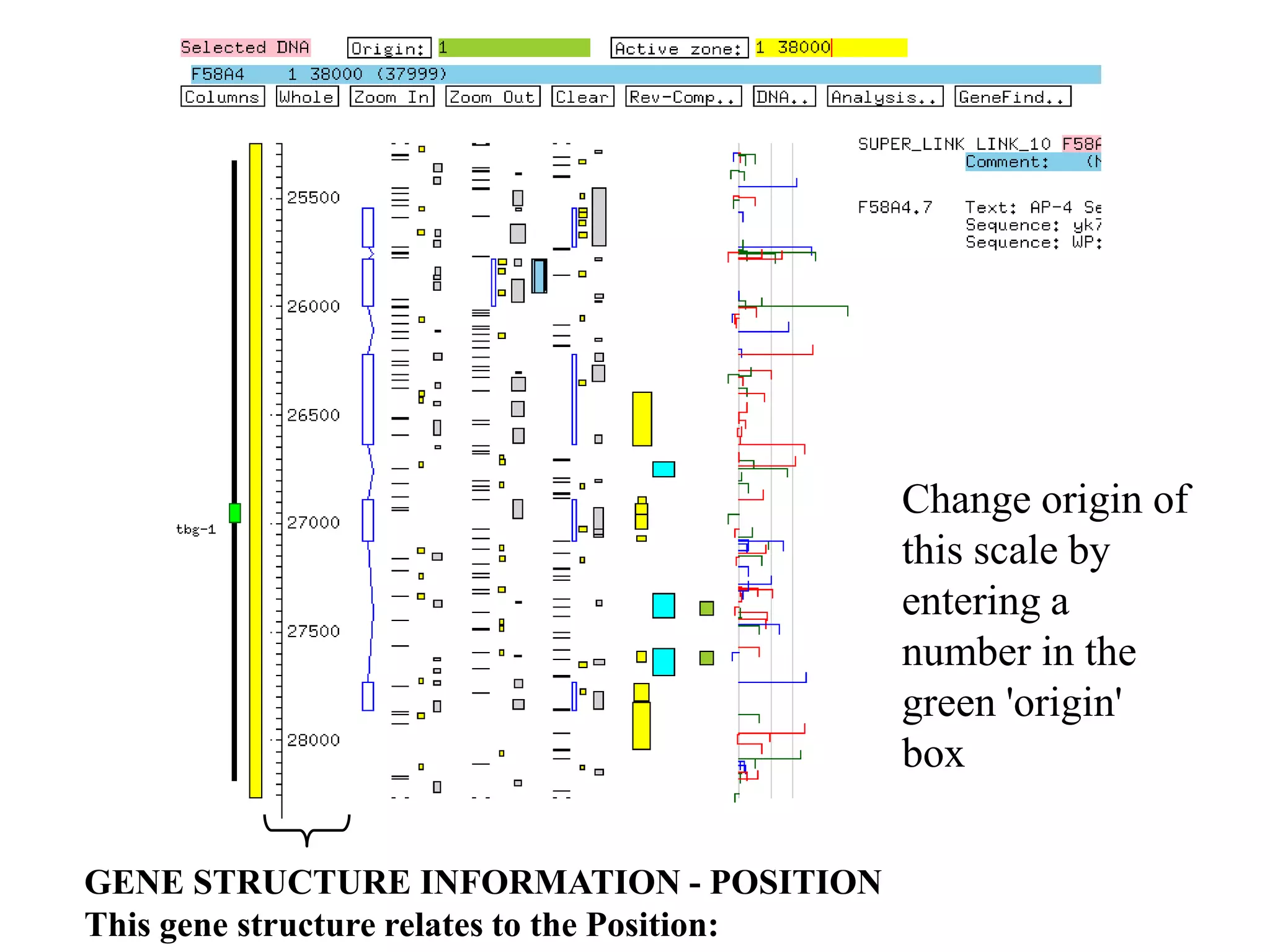
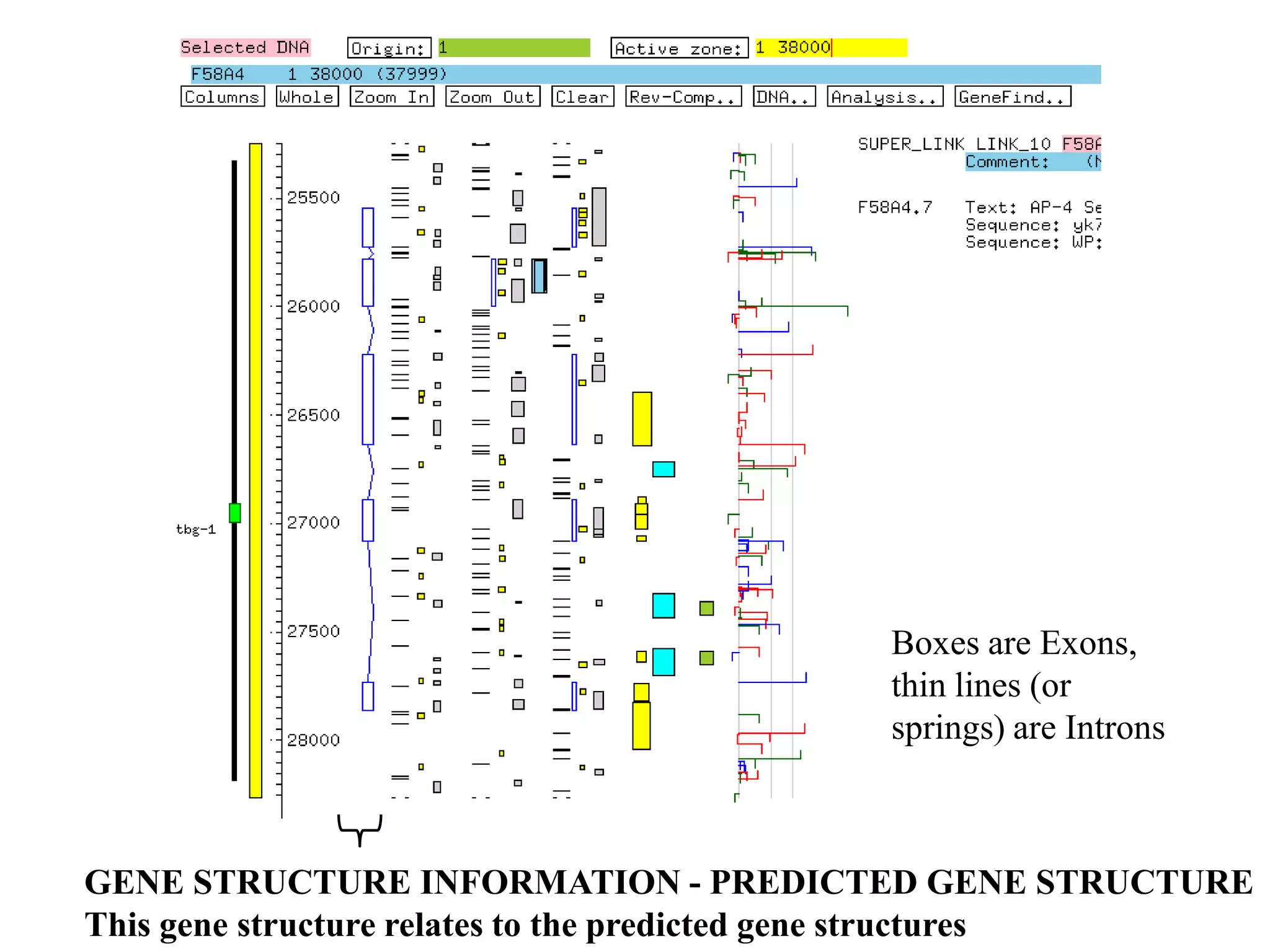
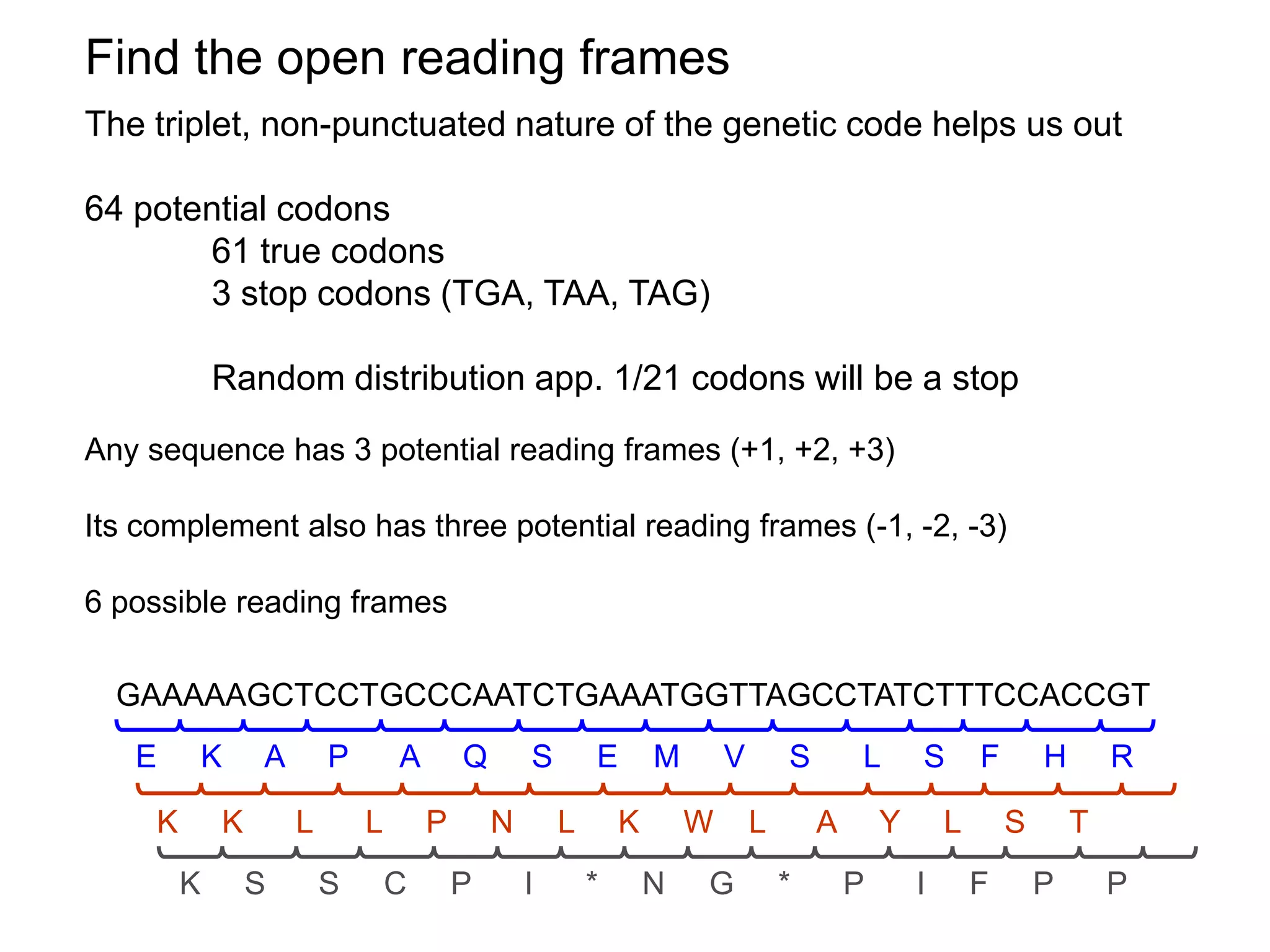
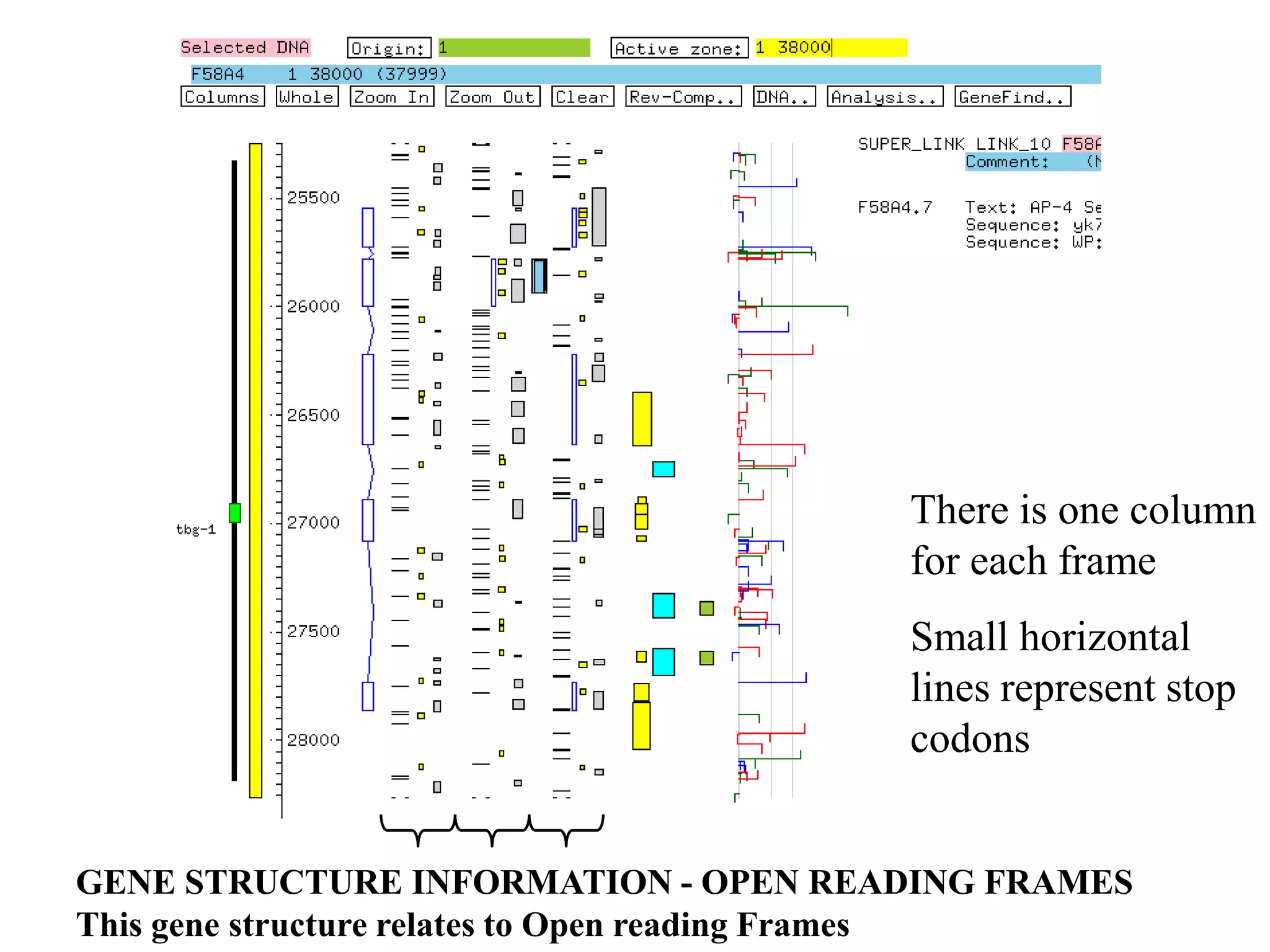
This document provides an overview of hidden Markov models (HMMs) and their application to gene prediction. It discusses how HMMs can model insertions and deletions in sequence alignments through their graphical representation using states and transitions. The document also explains how HMMs assign probabilities to sequences based on allowed state emissions and transitions. HMMs allow for more flexible modeling of gapped alignments than profiles or patterns alone.



![PATTERNS
• Small, highly conserved regions
• Shown as regular expressions
Example:
[AG]-x-V-x(2)-x-{YW}
– [] shows either amino acid
– X is any amino acid
– X(2) any amino acid in the next 2 positions
– {} shows any amino acid except these
BUT- limited to near exact match in small
region](https://image.slidesharecdn.com/bioinformatica-t8-go-hmm-121202135449-phpapp01/75/Bioinformatica-t8-go-hmm-10-2048.jpg)




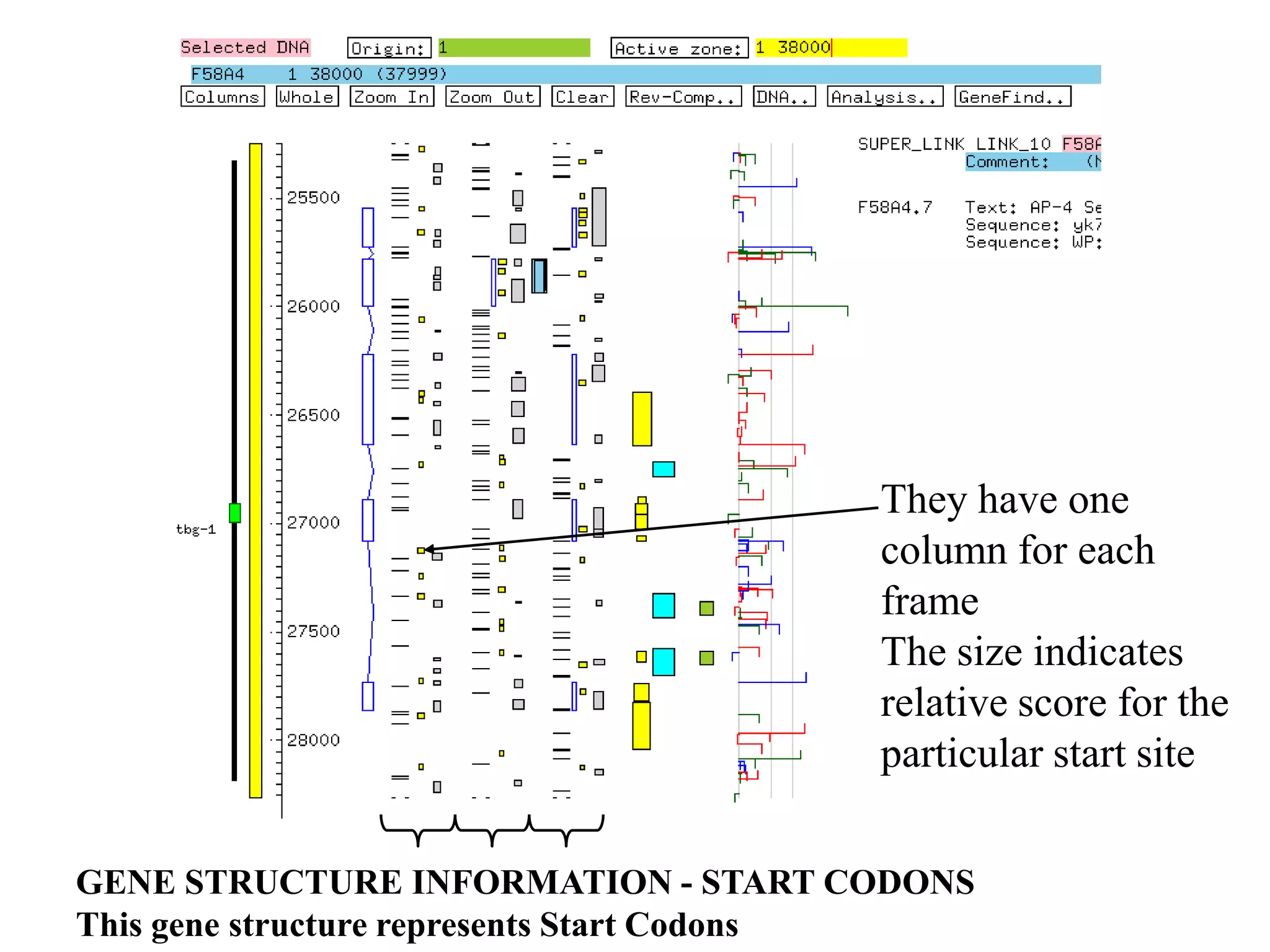
![Computational Gene Finding: Hexanucleotide frequencies
• Amino acid distributions are biased
e.g. p(A) > p(C)
• Pairwise distributions also biased
e.g. p(AT)/[p(A)*p(T)] > p(AC)/[p(A)*p(C)]
• Nucleotides that code for preferred amino
acids (and AA pairs) occur more frequently in
coding regions than in non-coding regions.
• Codon biases (per amino acid)
• Hexanucleotide distributions that reflect those
biases indicate coding regions.](https://image.slidesharecdn.com/bioinformatica-t8-go-hmm-121202135449-phpapp01/75/Bioinformatica-t8-go-hmm-38-2048.jpg)