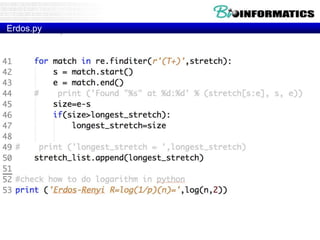
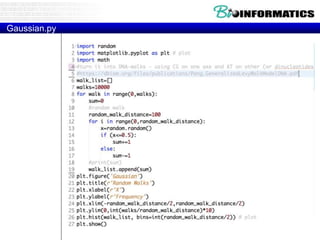
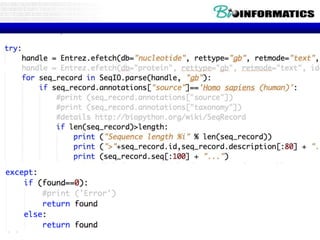
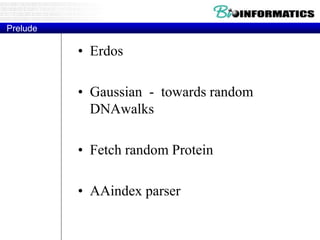
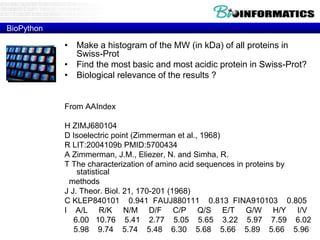
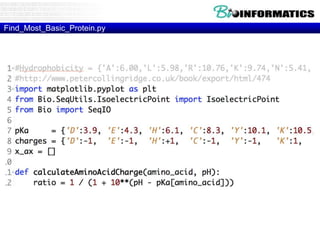
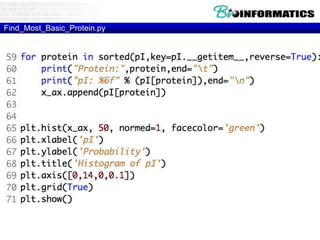
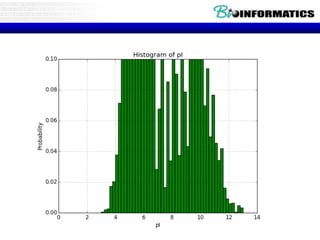
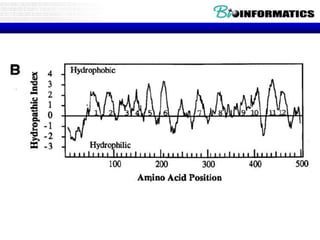
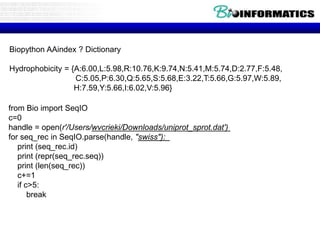
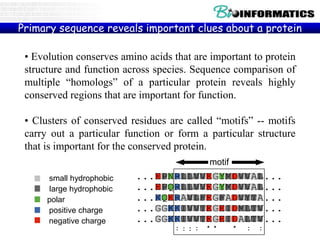
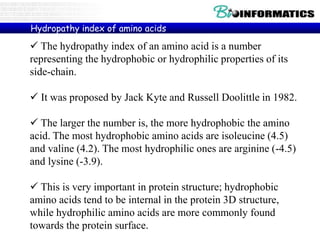
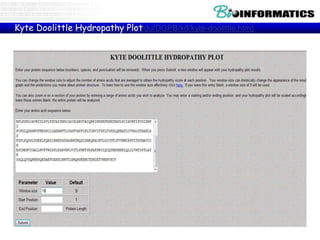
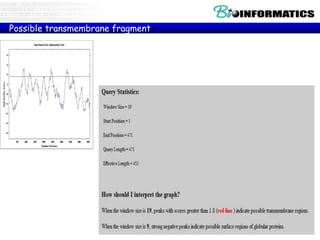
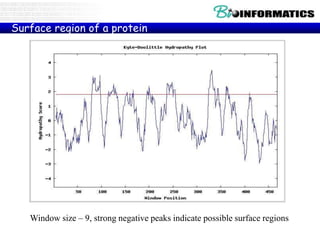
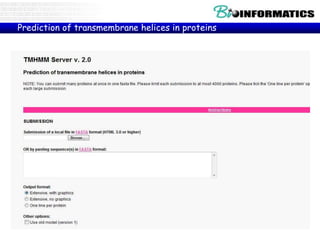
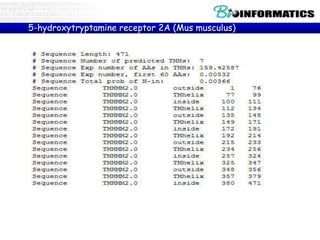
The document provides information on various Python programming concepts including control structures, lists, dictionaries, regular expressions, exceptions, and biological applications using Biopython. It discusses if/else statements, while and for loops, list operations, dictionary usage, regex patterns, exception handling roles, and gives examples analyzing protein sequences and structures using Biopython.



![Control Structures
if condition:
statements
[elif condition:
statements] ...
else:
statements
while condition:
statements
for var in sequence:
statements
break
continue](https://image.slidesharecdn.com/p62017biopython2-171127220435/85/P6-2017-biopython2-5-320.jpg)
![Lists
• Flexible arrays, not Lisp-like linked
lists
• a = [99, "bottles of beer", ["on", "the",
"wall"]]
• Same operators as for strings
• a+b, a*3, a[0], a[-1], a[1:], len(a)
• Item and slice assignment
• a[0] = 98
• a[1:2] = ["bottles", "of", "beer"]
-> [98, "bottles", "of", "beer", ["on", "the", "wall"]]
• del a[-1] # -> [98, "bottles", "of", "beer"]](https://image.slidesharecdn.com/p62017biopython2-171127220435/85/P6-2017-biopython2-6-320.jpg)
![Dictionaries
• Hash tables, "associative arrays"
• d = {"duck": "eend", "water": "water"}
• Lookup:
• d["duck"] -> "eend"
• d["back"] # raises KeyError exception
• Delete, insert, overwrite:
• del d["water"] # {"duck": "eend", "back": "rug"}
• d["back"] = "rug" # {"duck": "eend", "back":
"rug"}
• d["duck"] = "duik" # {"duck": "duik", "back":
"rug"}](https://image.slidesharecdn.com/p62017biopython2-171127220435/85/P6-2017-biopython2-7-320.jpg)
![Regex.py
text = 'abbaaabbbbaaaaa'
pattern = 'ab'
for match in re.finditer(pattern, text):
s = match.start()
e = match.end()
print ('Found "%s" at %d:%d' % (text[s:e], s, e))
m = re.search("^([A-Z]) ",line)
if m:
from_letter = m.groups()[0]](https://image.slidesharecdn.com/p62017biopython2-171127220435/85/P6-2017-biopython2-8-320.jpg)