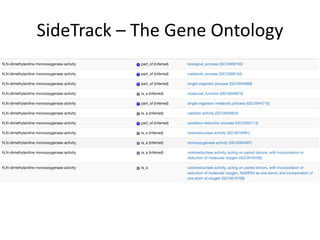
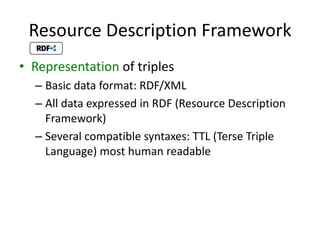
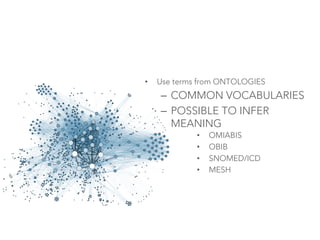
This document provides an introduction to bio-ontologies and the semantic web. It discusses what ontologies are and how they are used in the bio domain through initiatives like the OBO Foundry. It introduces key semantic web technologies like RDF, URIs, Turtle syntax, and SPARQL query language. It provides examples of ontologies like the Gene Ontology and how ontologies can be represented and queried using these semantic web standards.






















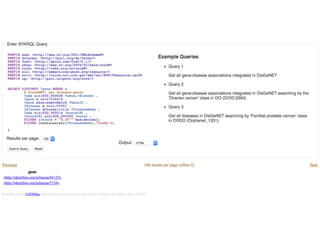
![The Turtle Syntax
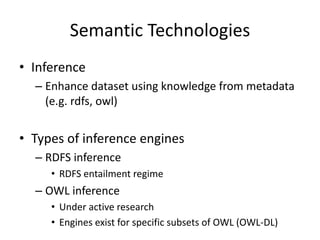
• Blank nodes
@prefix b4x: <http:bioinformatics.be/terms#> .
@prefix foaf: <http://xmlns.com/foaf/0.1/> .
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .
b4x:martijn b4x:has_favorite_beer b4x:karmeliet,
b4x:chimay_blauw;
b4x:length “184”^^xsd:integer;
foaf:name “Martijn Devisscher”^^xsd:string;
b4x:owns_cat [ b4x:color “Gray” ].](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-25-320.jpg)













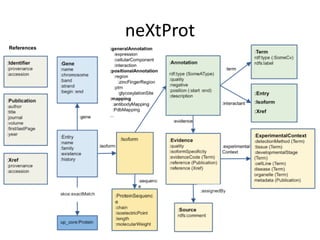
![RDFS: Inference
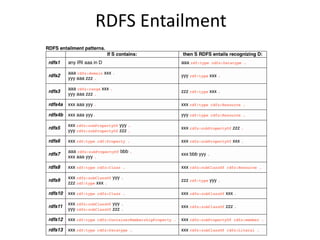
b4x:kevin b4x:has_favorite_beer b4x:stella
Inferred triples:
b4x:kevin a foaf:Person [from domain]
b4x:stella a b4x:Beer [from range]
b4x:stella a b4x:Drink [from subClassOf]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-43-320.jpg)








































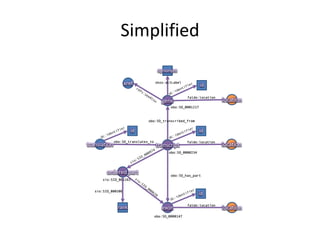

















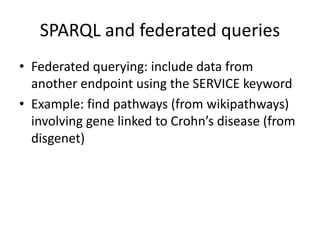
![PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX void: <http://rdfs.org/ns/void#>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#>
PREFIX up: <http://purl.uniprot.org/core/>
SELECT DISTINCT ?disease WHERE {
?gda a [(rdfs:subClassOf)* sio:SIO_000983].
?gda sio:SIO_000628 ?disease.
?disease a ncit:C7057.
?gda sio:SIO_000628 ?gene.
?gene a ncit:C16612.
?gene skos:exactMatch <http://identifiers.org/hgnc.symbol/BRCA1>}
http://rdf.disgenet.org/lodestar/sparql](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-108-320.jpg)