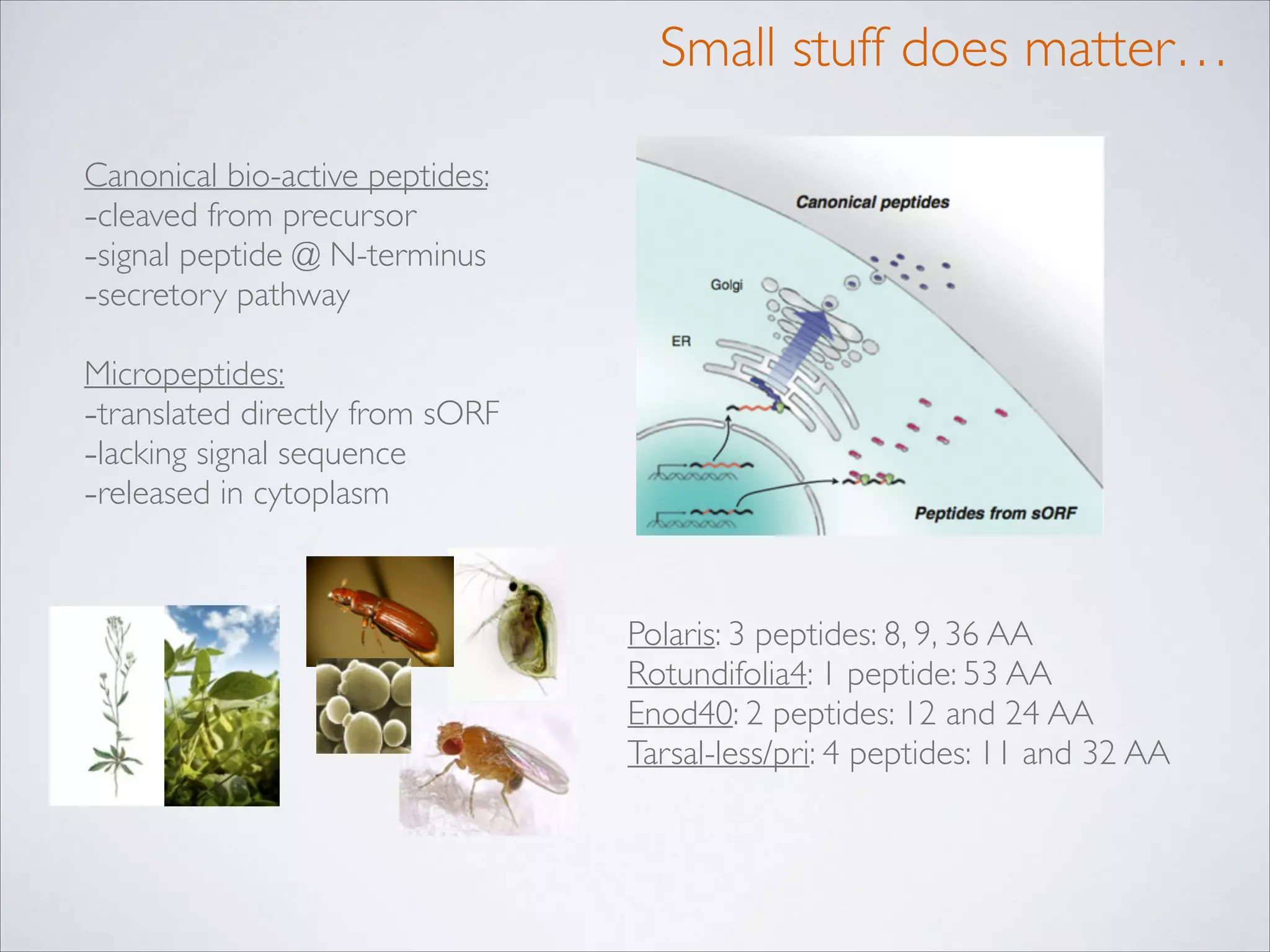
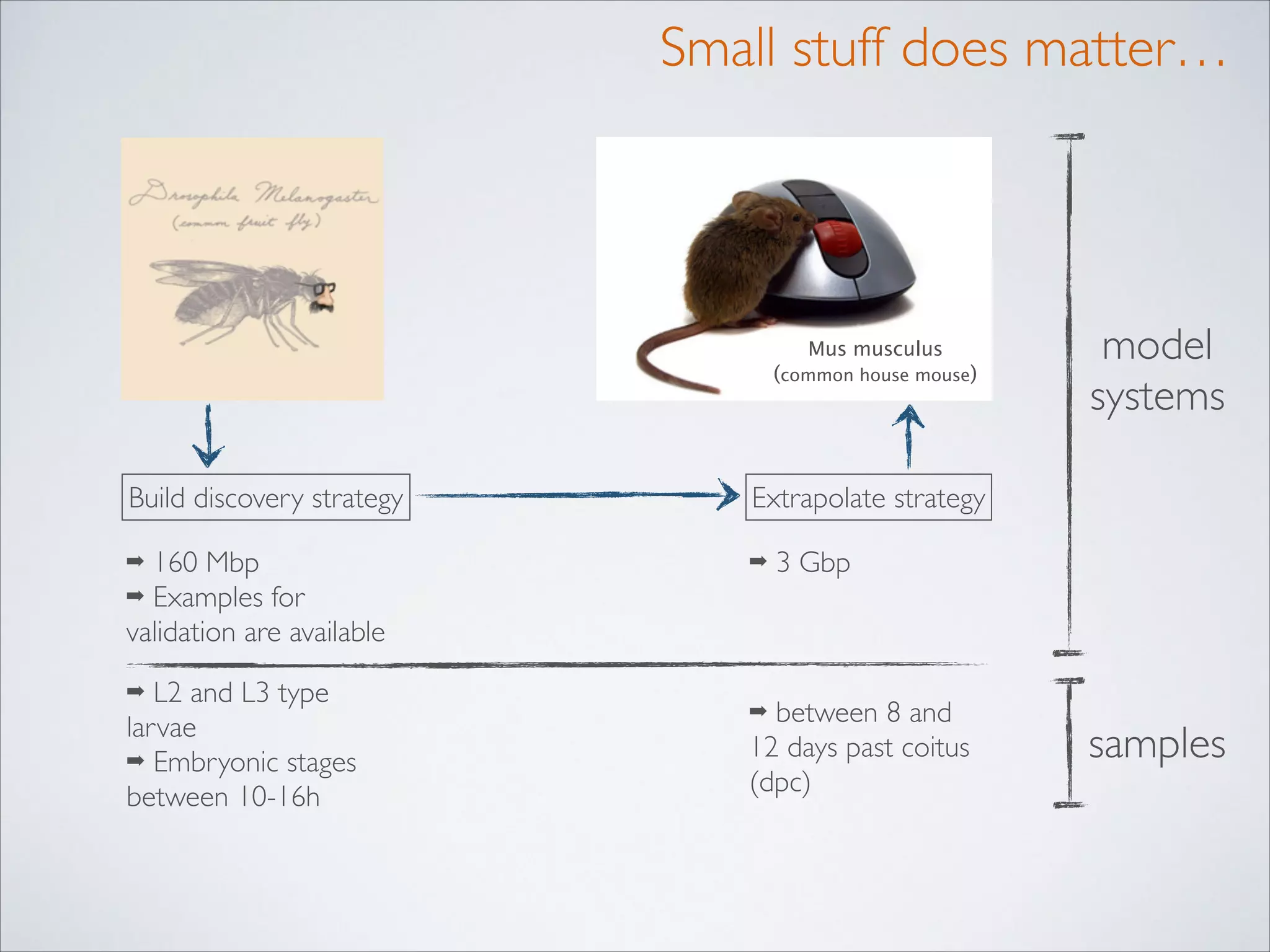
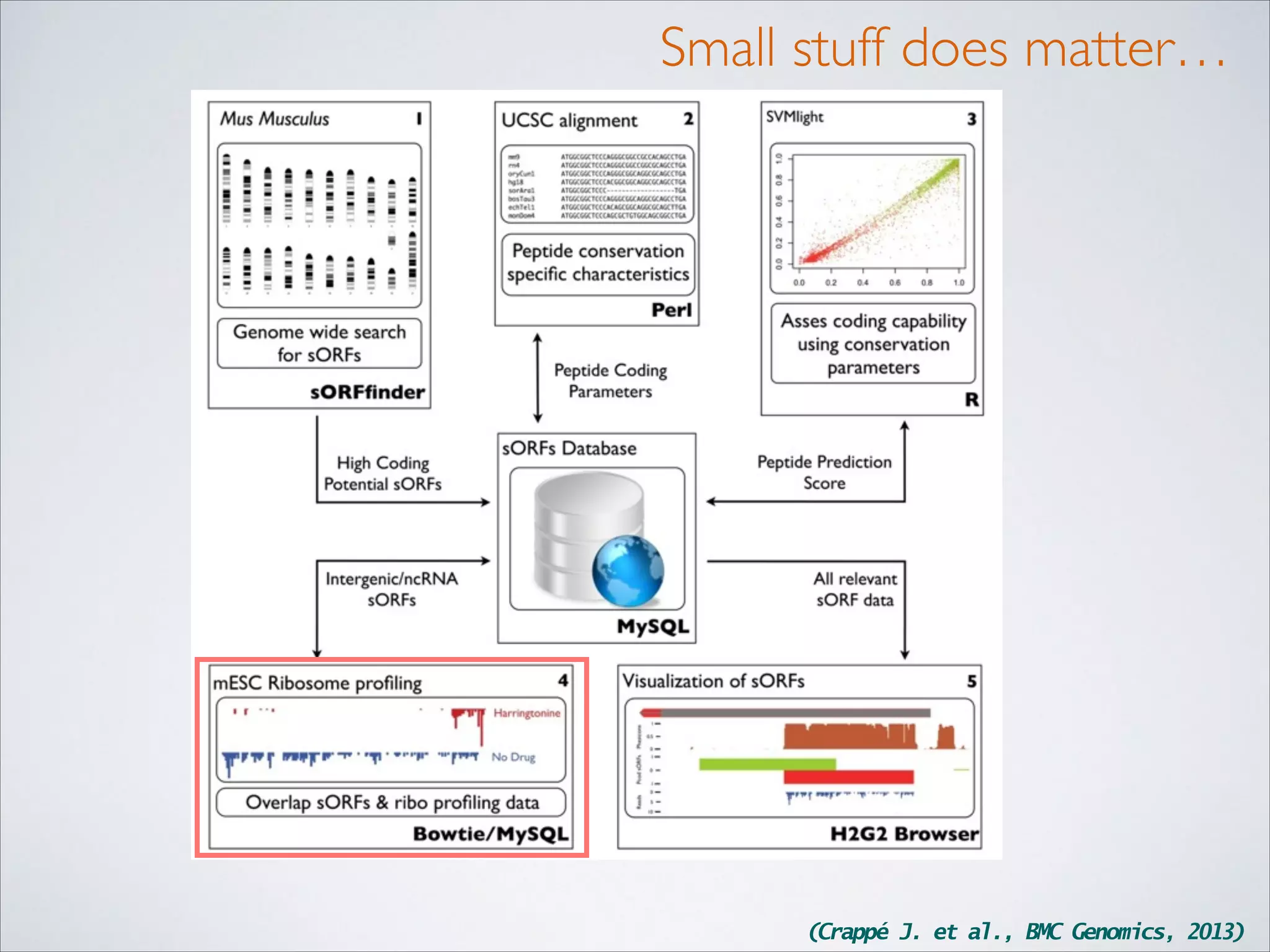
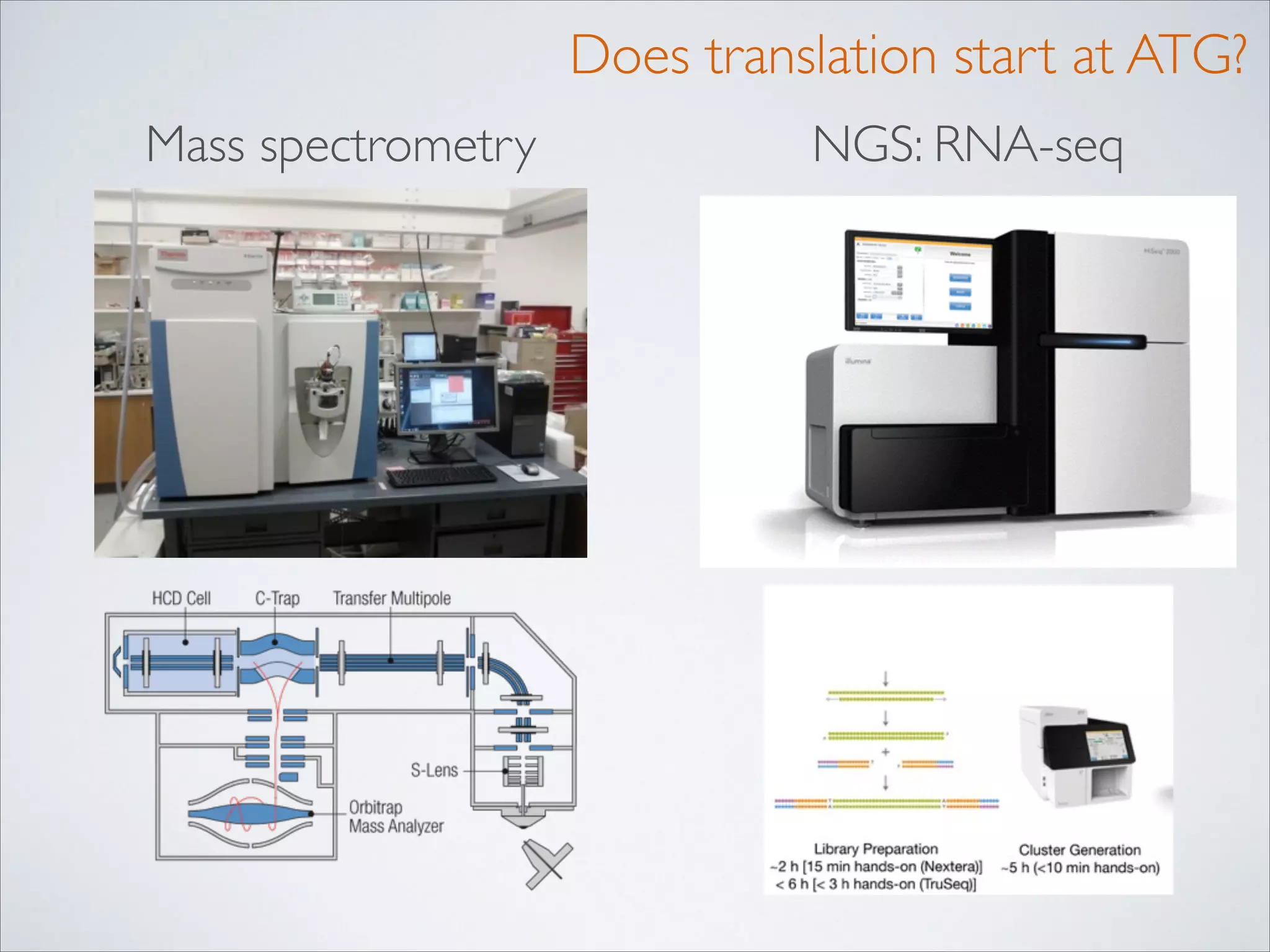
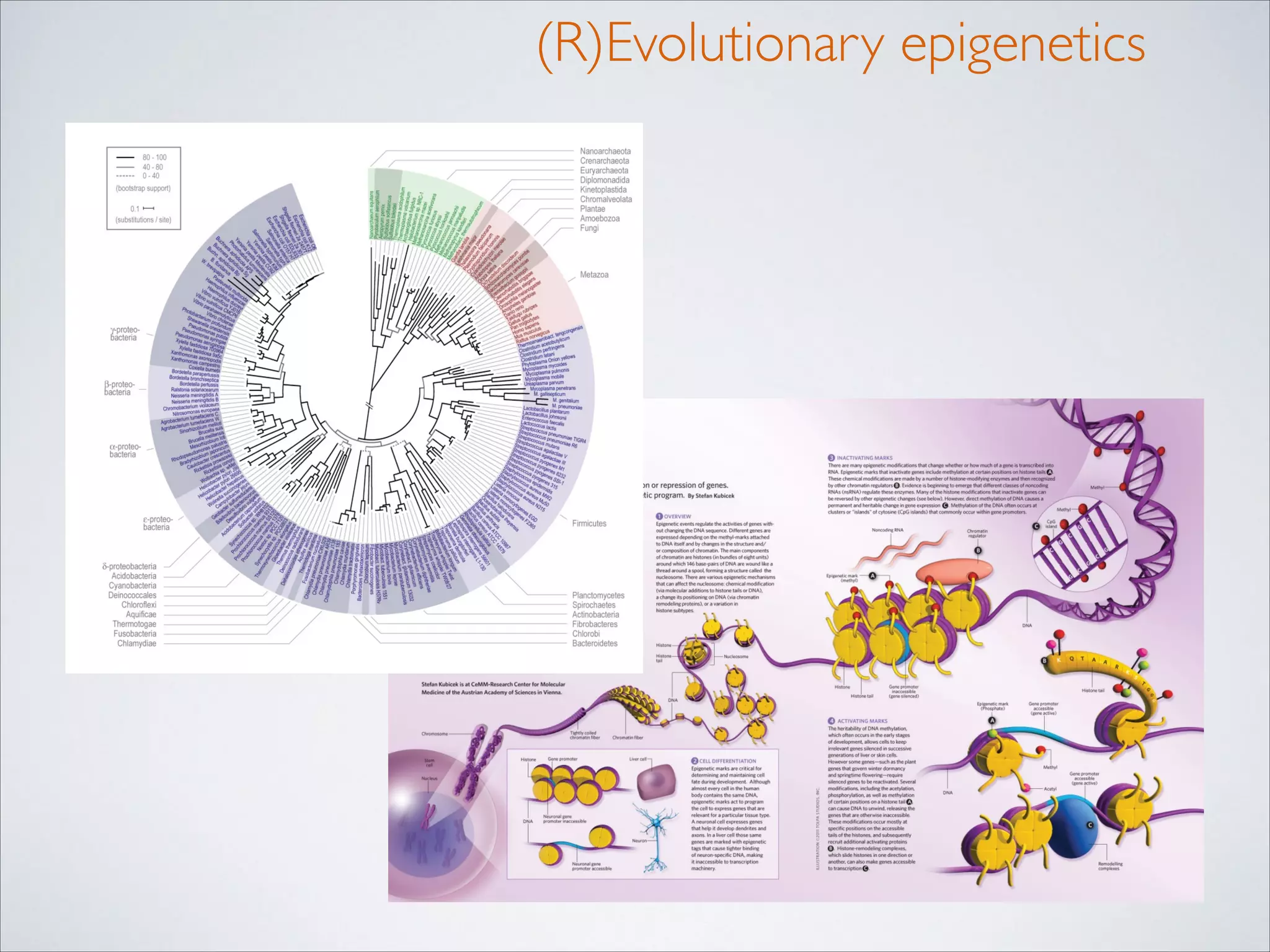
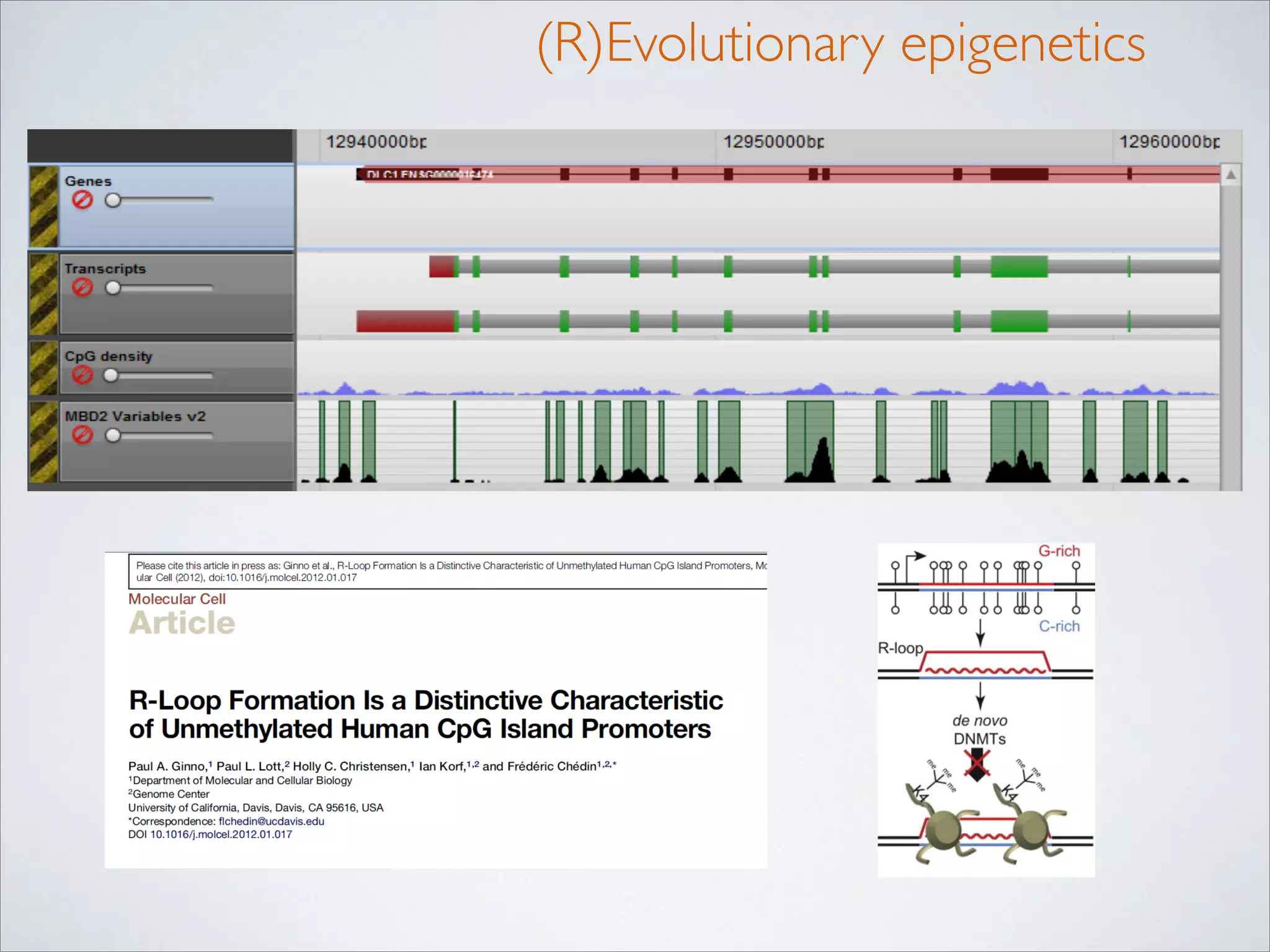
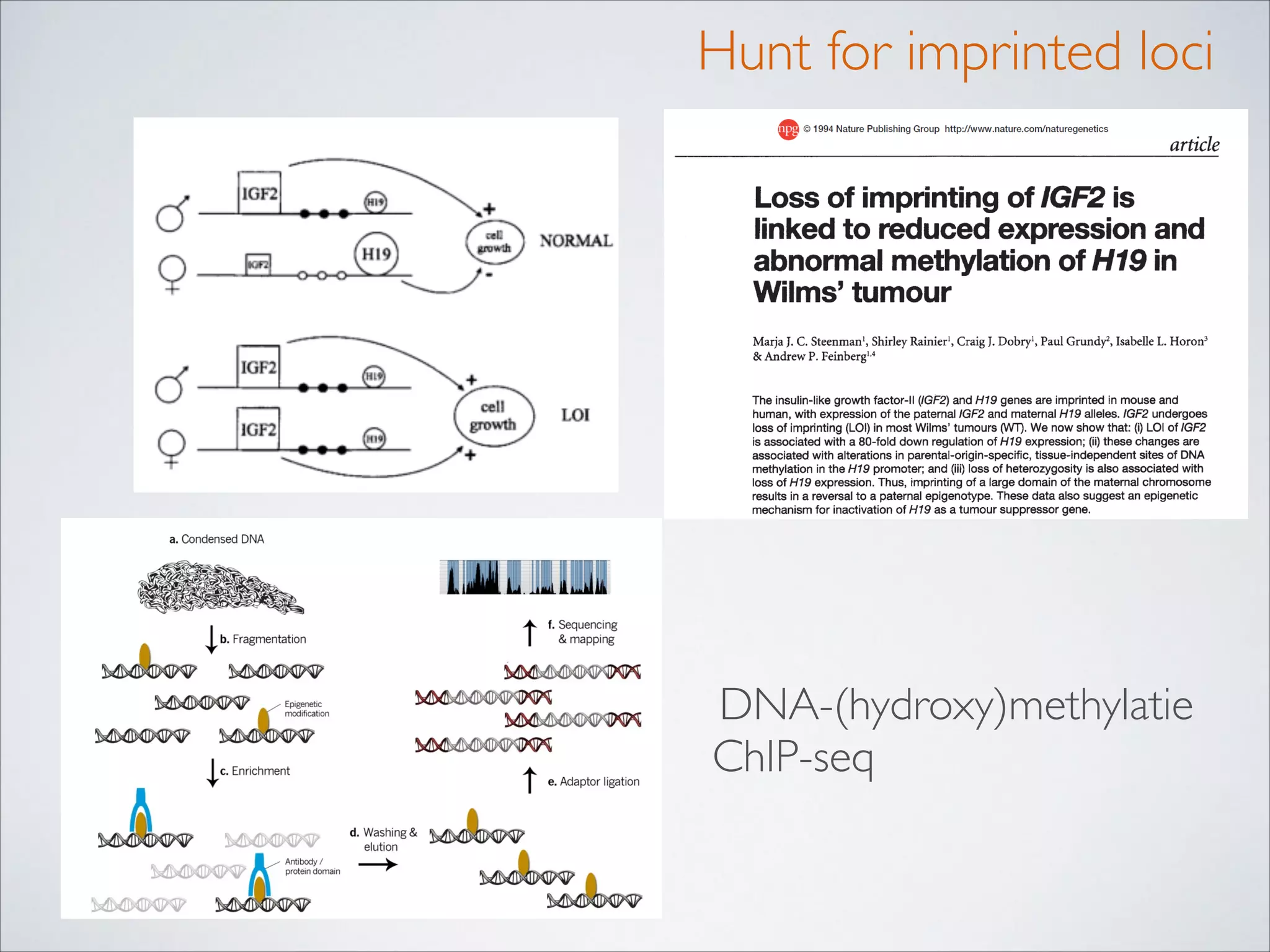
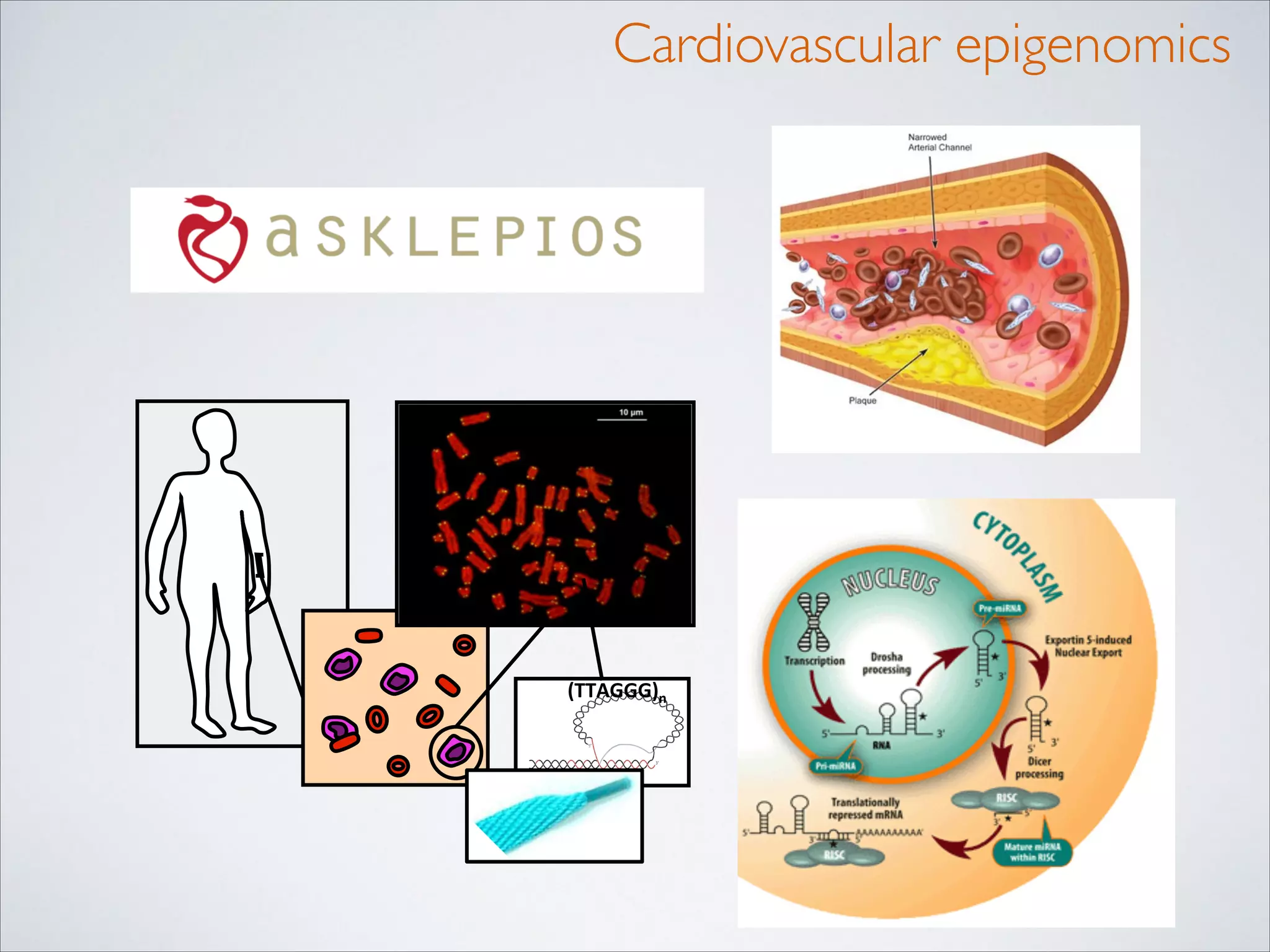
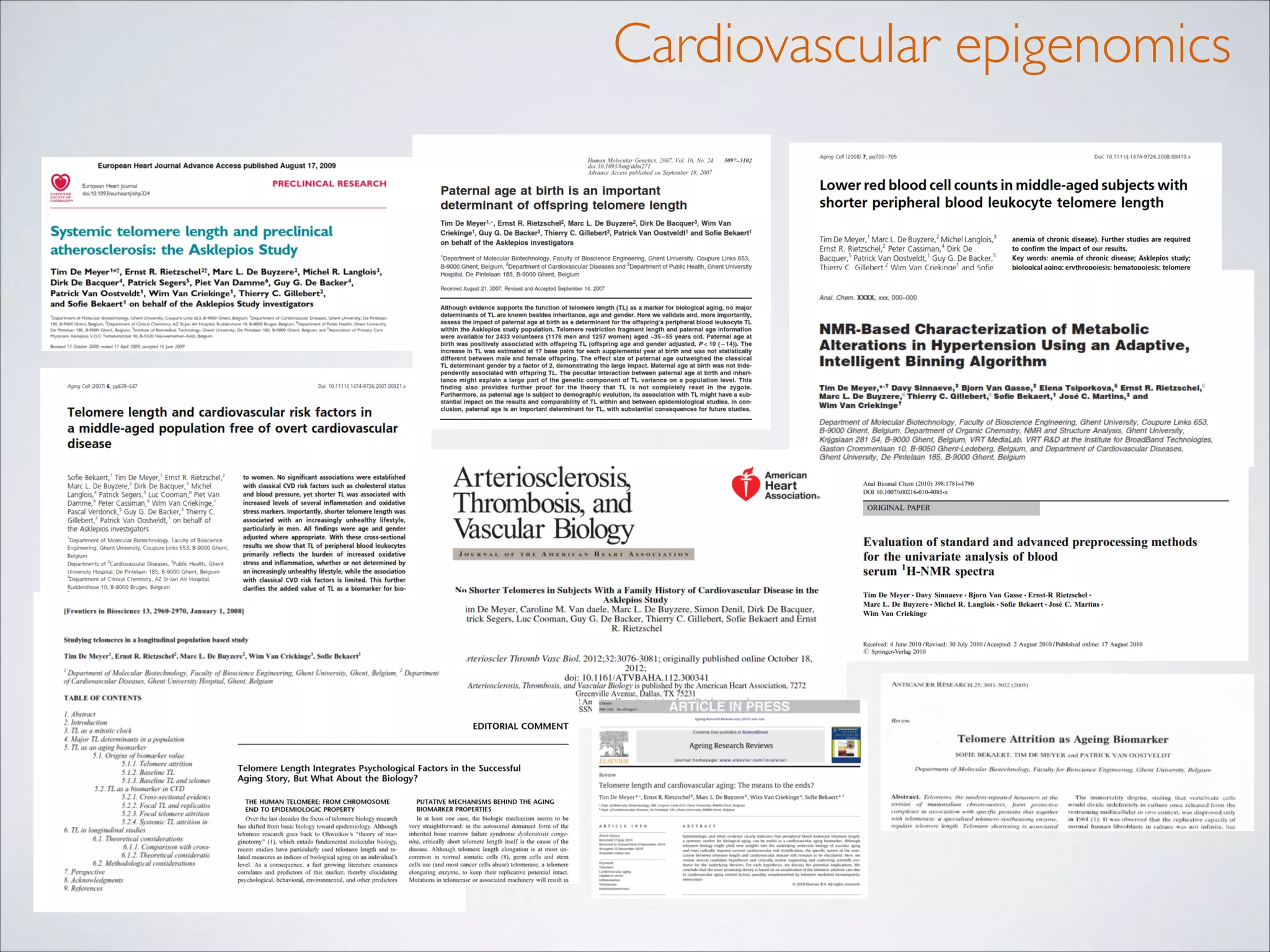
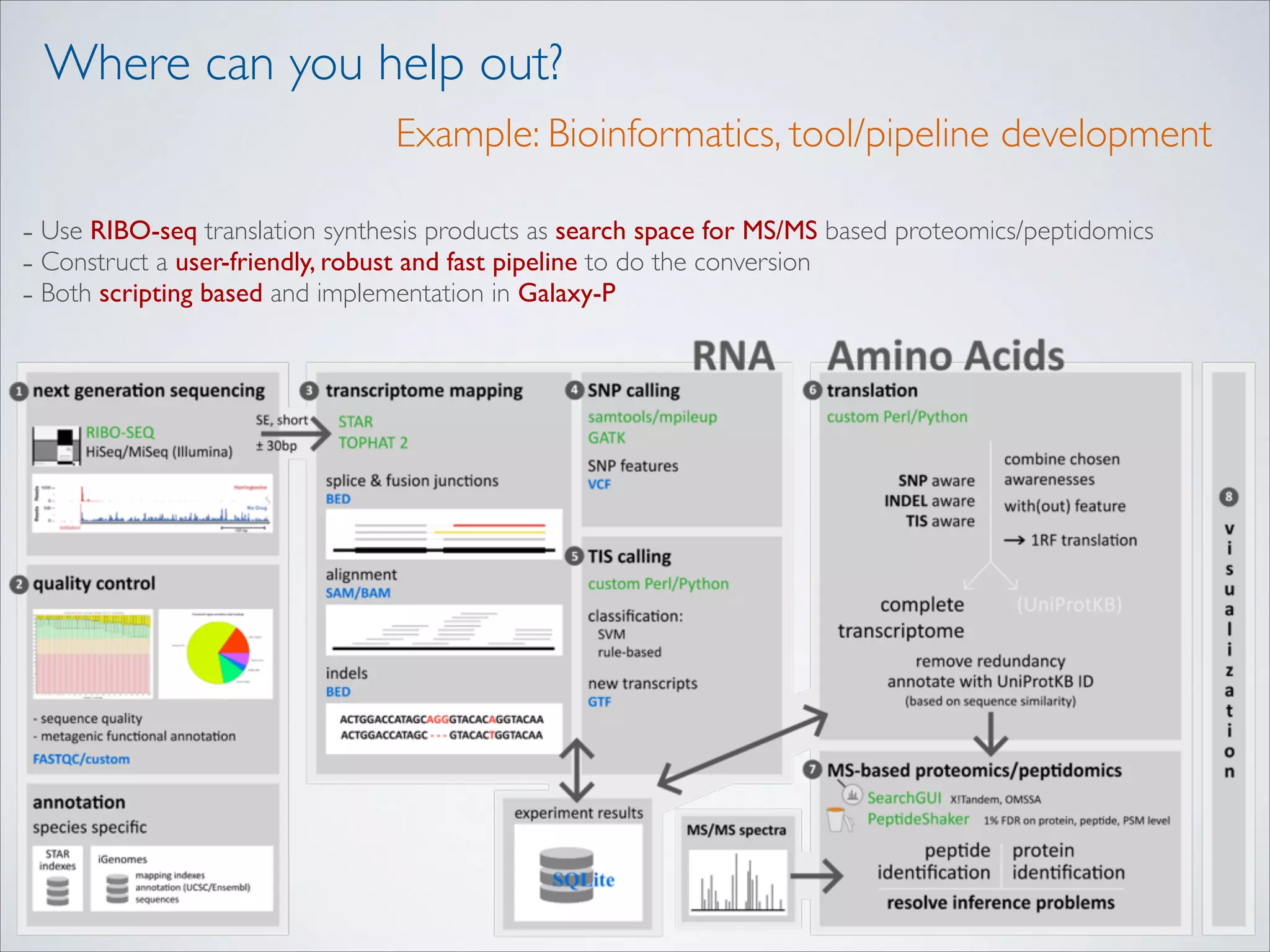
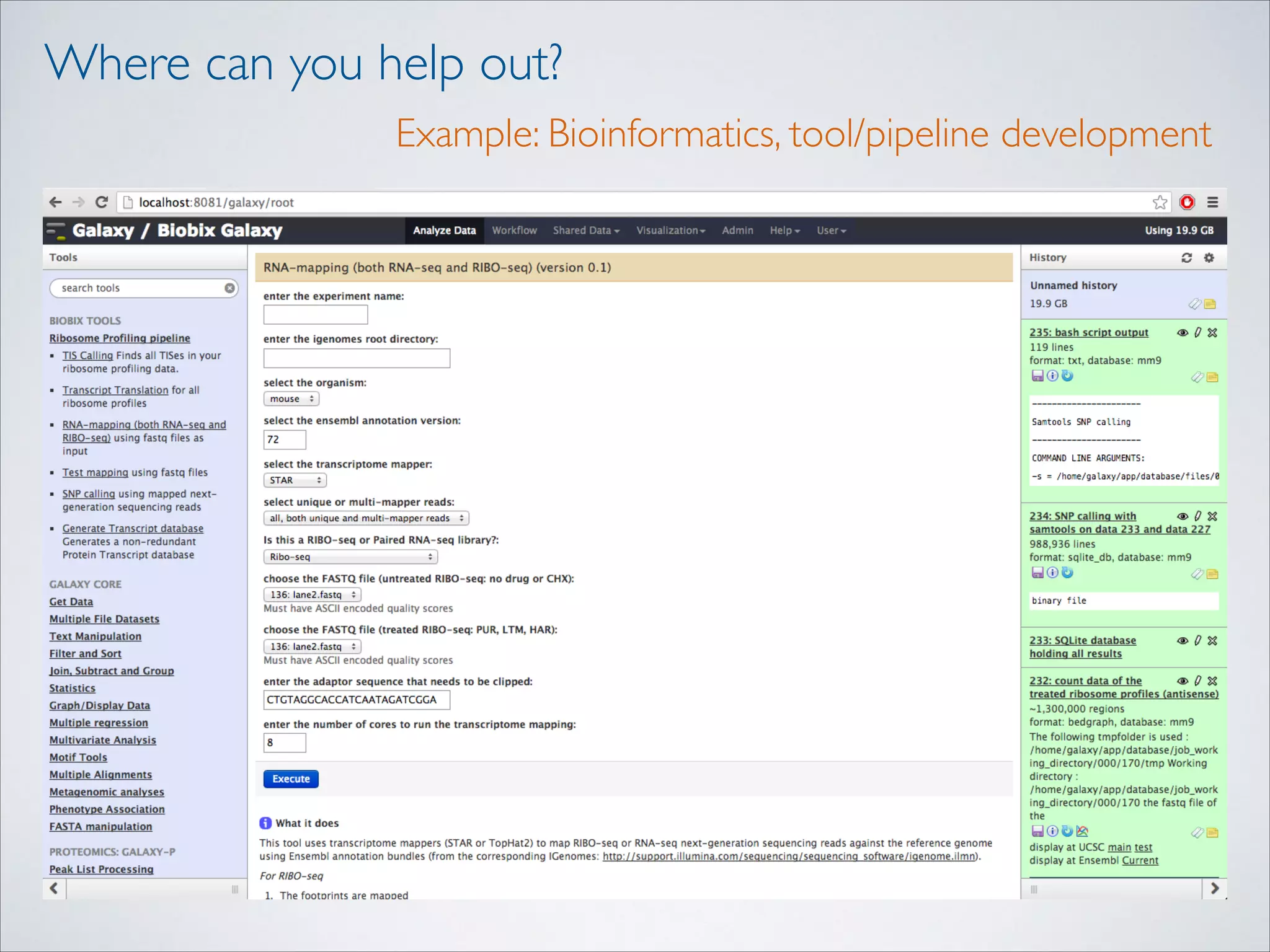
This master's thesis project involves studying small regulatory peptides and alternative translation initiation sites using techniques like ribosome profiling, mass spectrometry, and sequencing. Specific topics being investigated include micropeptides, non-ATG translation start sites, nuclear translation, evolutionary epigenetics, imprinted loci identification, and cardiovascular epigenomics. The student is welcome to contribute to tool development for analyzing ribosome profiling and mass spectrometry data, or help with computational analysis and data integration. Potential collaborators mentioned include labs at VIB, VITO, and international institutions studying related topics.



![Example: Bioinformatics, tool/pipeline development
(3) Start codon: both cognate and near-cognate
1.0
ACG
0.0
G
TG
G
C
A
A
C
G
C
T
TA
C
G
T
1.0
probability
bits
2.0
T
0.5
G
TG
GA
ACCG
A
GTAC
C
0.0 TGT TC
T
C
A
5
Kozak
mo(f:[A/G]CCatgG[not
T]
Based on new N-term-ext and uORF identifications
T
C
A
5
WebLogo 3.3
WebLogo 3.3
(4) Example: HDGF_MOUSE (n-term-ext, near-cognate start site)
GTG
GTG
TTG
HDGF
(hsa)
stomach&
skin&
spleen&
heart&
kidney&
intes*ne&
lung&
46#
40#kDa#
38#kDa#
36.5#kDa#
HDGF
(mmu)
(V)AAPELASGAGIEAGAAR
(|)||||| || |||||||
(V)AAPELGPGATIEAGAAR
liver&
tes*s&
GCCGTGTGTTGCCCACCGCGCCCGGCCCTGTCCGA
GCGGCGCGCGGGCGCAGACGCCGTGGCTGCCCCGG
AGCTCGCGTCGGGGGCCGGCATCGAGGCGGGGGCC
GCGCGAGGGCCGGAGCGCAGCGGCGCCGCAACCGC
CGCACGCGCAAACTTGGGCTCGCGCTTCCCGGCTC
GGCGCGGAGCCCGGGGCGCCCGCGGCCCCGCCATG
TCGCGATCCAACCGGCAGA…
brain&
uc008ptc.1
muscle&
ATG
32#
HDGF%
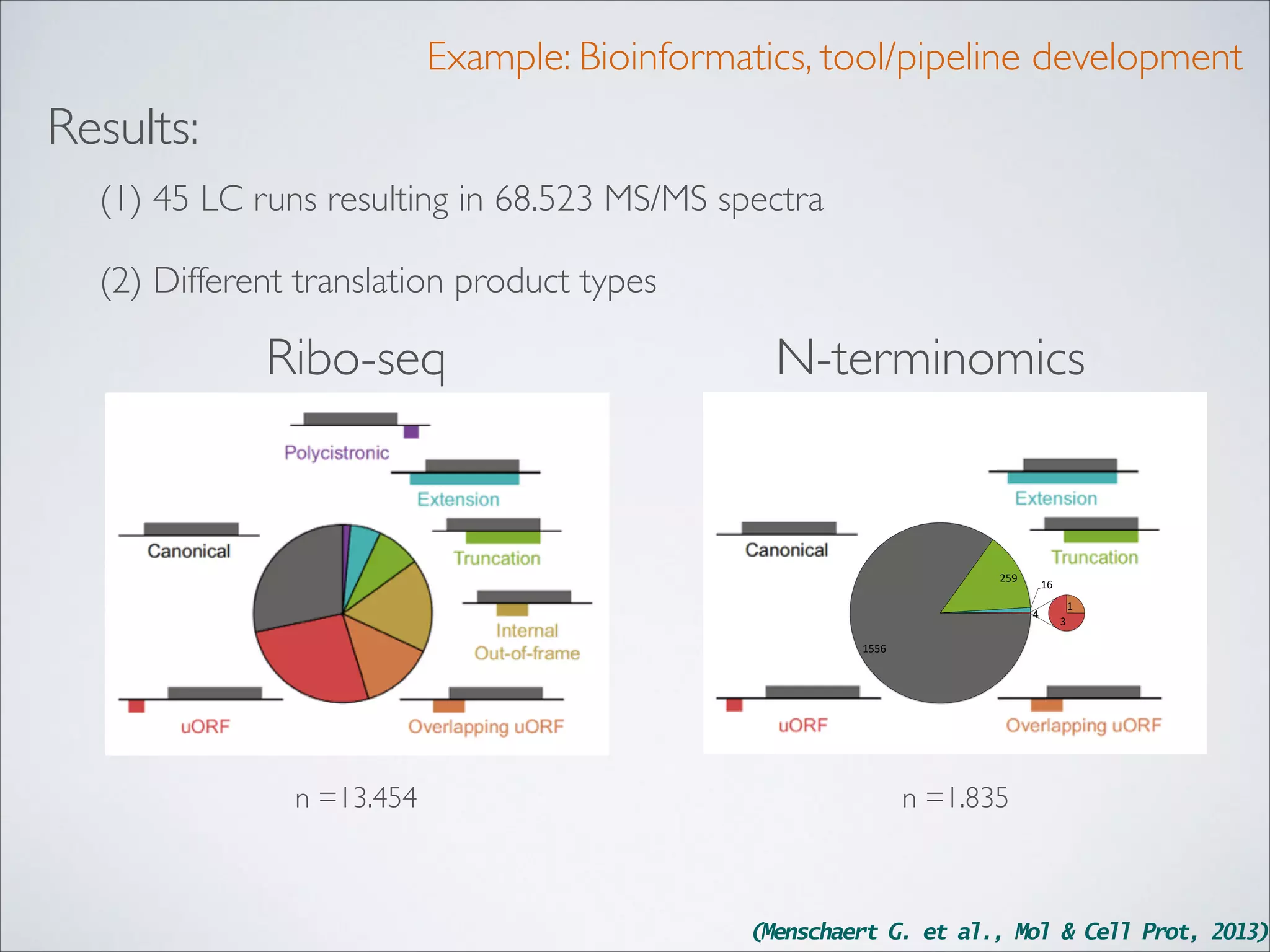
(Menschaert G. et al., Mol & Cell Prot, 2013)](https://image.slidesharecdn.com/thesisbiobix-131105143638-phpapp02/75/Thesis-biobix-20-2048.jpg)

