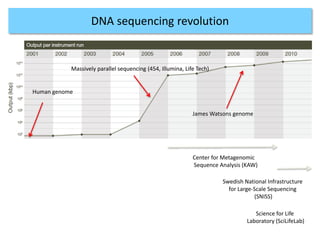
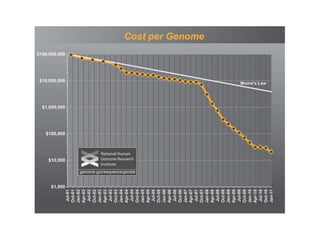
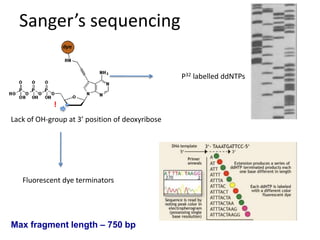
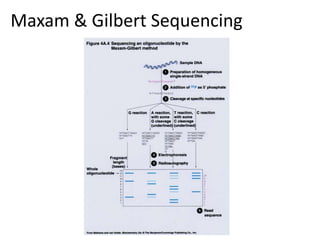
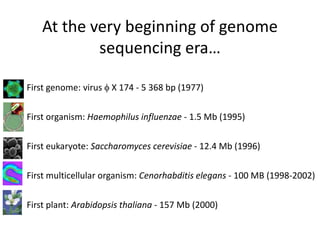
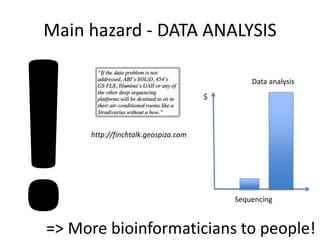
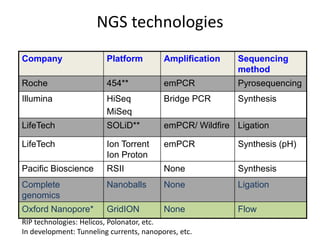
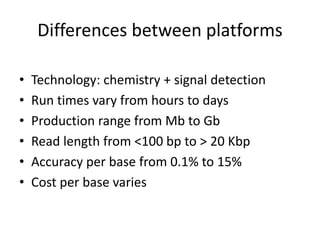
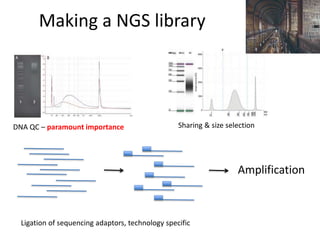
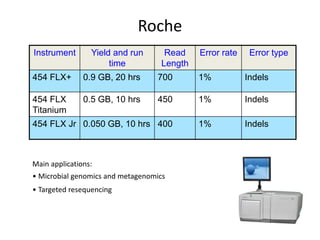
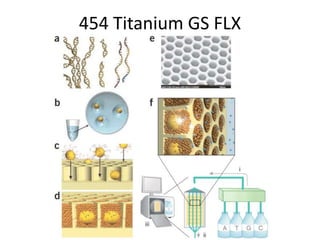
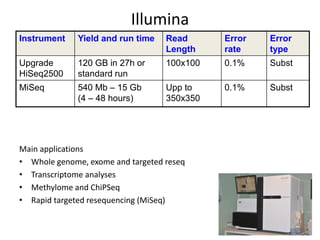
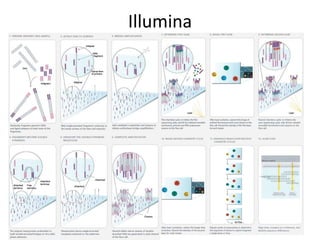
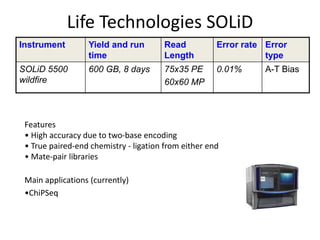
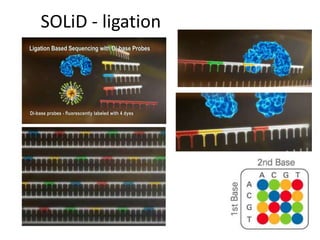
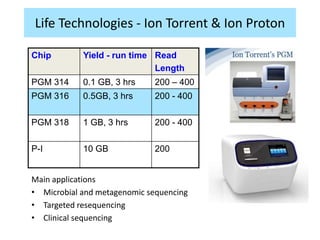
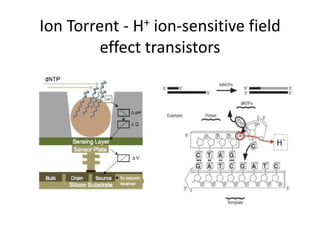
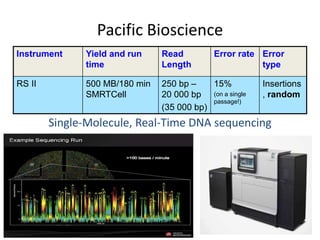
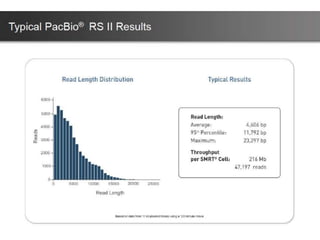
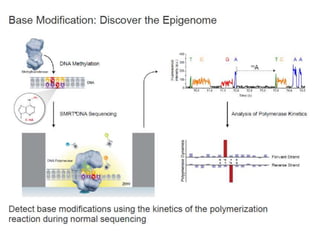
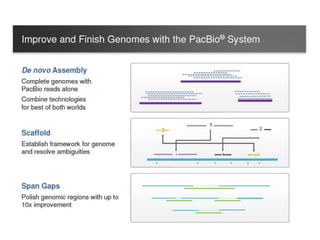
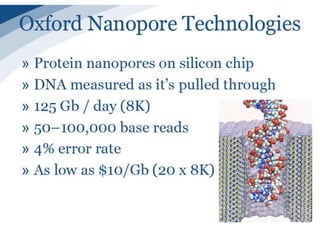
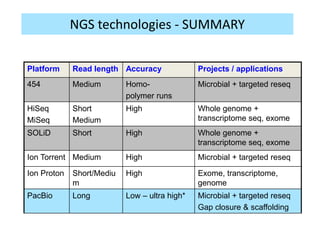
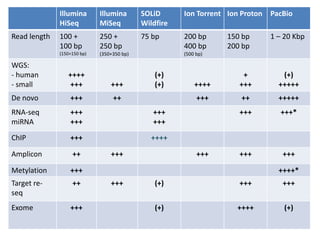
The document discusses genomic research and sequencing technologies. It provides a history of genomic research from early sequencing methods like Sanger sequencing to modern massively parallel sequencing. It describes several next-generation sequencing platforms, including their read lengths, accuracy, applications, and differences. It emphasizes that data analysis is a major challenge and advises consulting sequencing facilities and having dedicated bioinformaticians for projects.