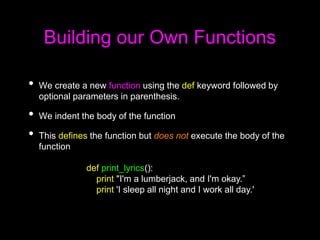
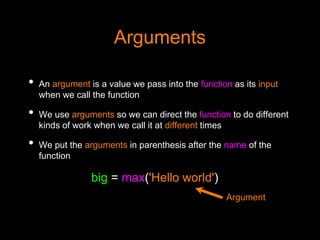
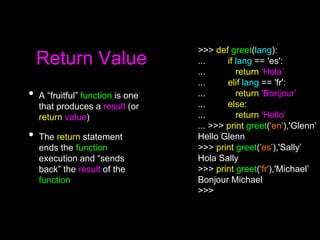
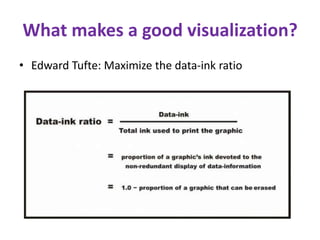
This document discusses Python functions. It explains that there are built-in functions provided as part of Python and user-defined functions. User-defined functions are created using the def keyword and can take parameters and return values. The body of a function is indented and runs when the function is called. Functions allow code to be reused and organized in a modular way. Examples are provided to demonstrate defining and calling functions with different parameters and return values.




![Recap
if condition:
statements
[elif condition:
statements] ...
else:
statements
while condition:
statements
for var in sequence:
statements
break
continue
Strings
REGULAR EXPRESSIONS](https://image.slidesharecdn.com/p42018iofunctions-181105221557/85/P4-2018-io_functions-5-320.jpg)






![import matplotlib.pyplot as plt
xs = range(-100,100,10)
x2 = [x**2 for x in xs]
negx2 = [-x**2 for x in xs]
plt.plot(xs, x2)
plt.plot(xs, negx2)
plt.xlabel("x”)
plt.ylabel("y”)
plt.ylim(-2000, 2000)
plt.axhline(0) # horiz line
plt.axvline(0) # vert line
plt.savefig(“quad.png”)
plt.show()
Incrementally
modify the figure.
Show it on the screen
Save your figure to a file](https://image.slidesharecdn.com/p42018iofunctions-181105221557/85/P4-2018-io_functions-15-320.jpg)









![Check installed libraries
import pip
import sys
import platform
import webbrowser
print ("Python " + platform.python_version()+ " installed packages:")
installed_packages = pip.get_installed_distributions()
installed_packages_list = sorted(["%s==%s" % (i.key, i.version)
for i in installed_packages])
print(*installed_packages_list,sep="n")](https://image.slidesharecdn.com/p42018iofunctions-181105221557/85/P4-2018-io_functions-26-320.jpg)
