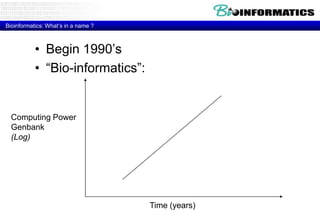
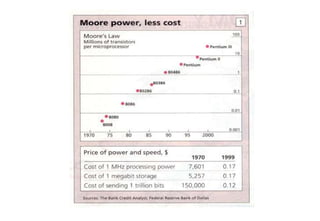
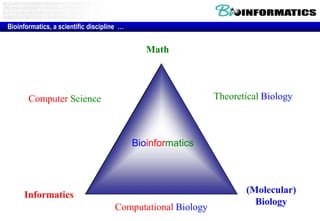
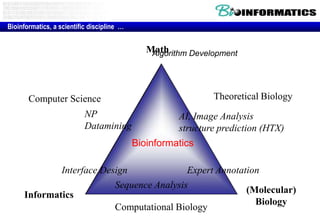
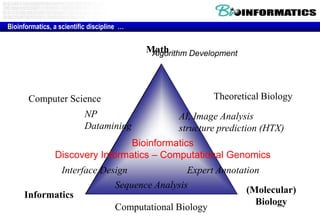
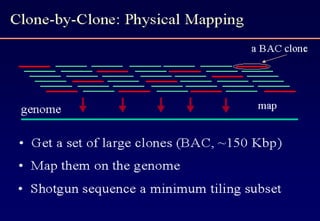
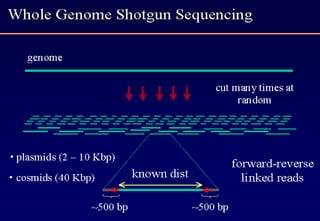
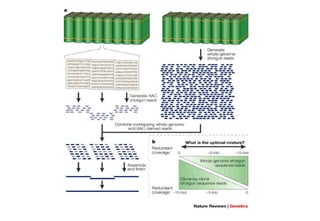
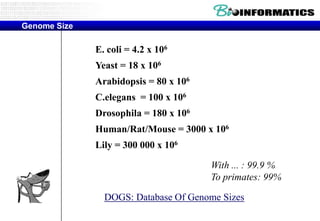
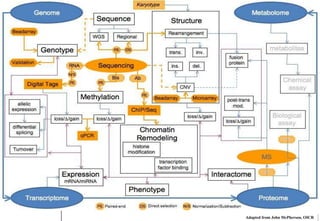
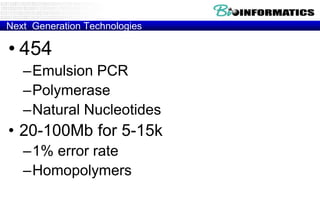
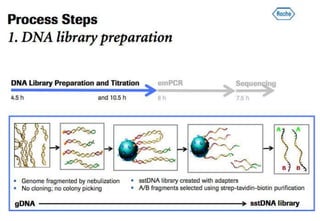
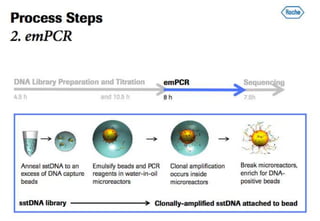
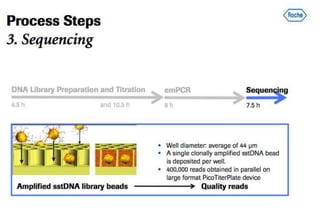
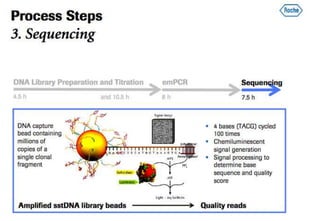
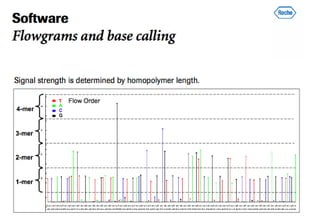
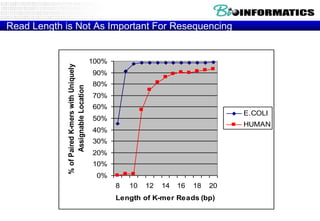
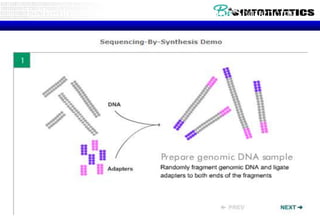
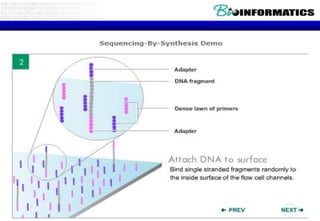
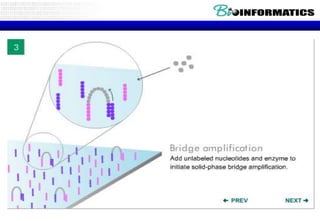
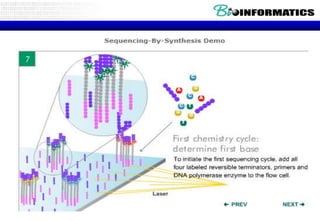
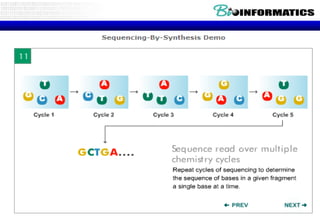
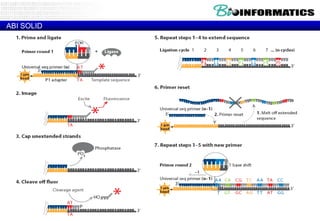
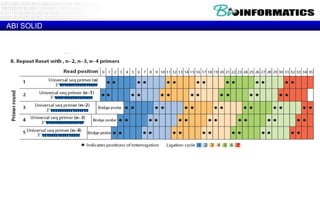
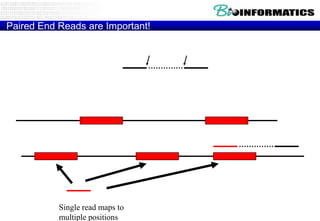
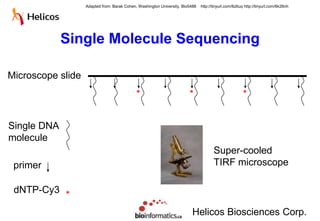
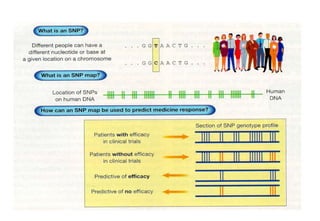
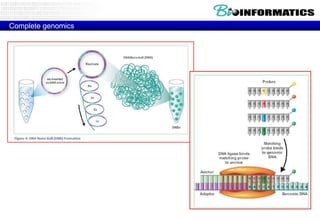
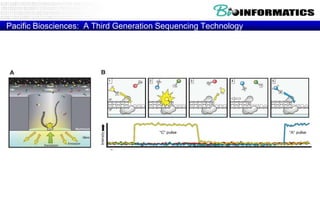
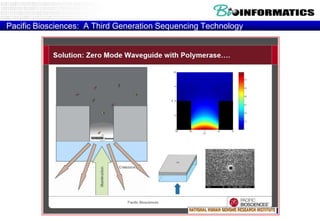
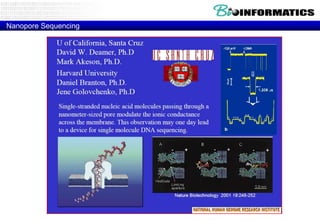
The document discusses the field of bioinformatics, which involves applying computational techniques and building tools to solve biological problems, such as analyzing genetic sequences and modeling molecular structures. It outlines several applications of bioinformatics, including in medicine for disease research and drug design, as well as in agriculture and animal health. The emergence of bioinformatics is attributed to the convergence of rapid growth in fields like biotechnology and information technology.