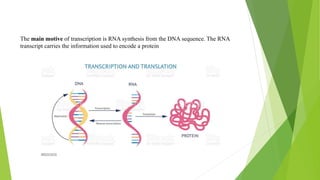
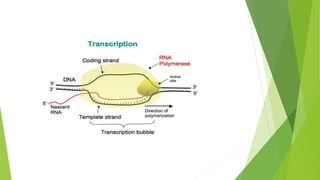
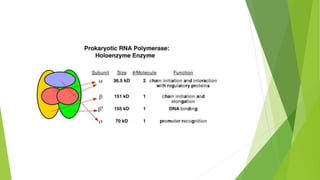
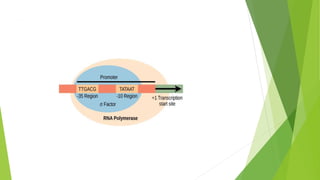
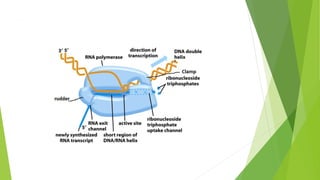
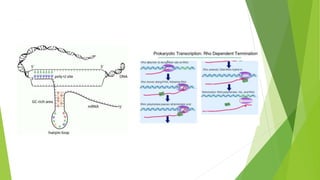
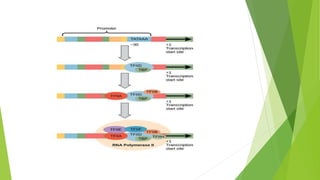
The document provides information about the process of transcription. It defines transcription as the first step of gene expression where an RNA molecule is formed from DNA. It describes the key steps and components of transcription in both prokaryotes and eukaryotes. These include initiation, elongation, and termination of transcription, as well as the enzymes like RNA polymerase that are involved. The summary highlights that transcription involves copying genetic information from DNA to RNA, and differs between single-celled and multi-cellular organisms in its regulation and RNA polymerases used.