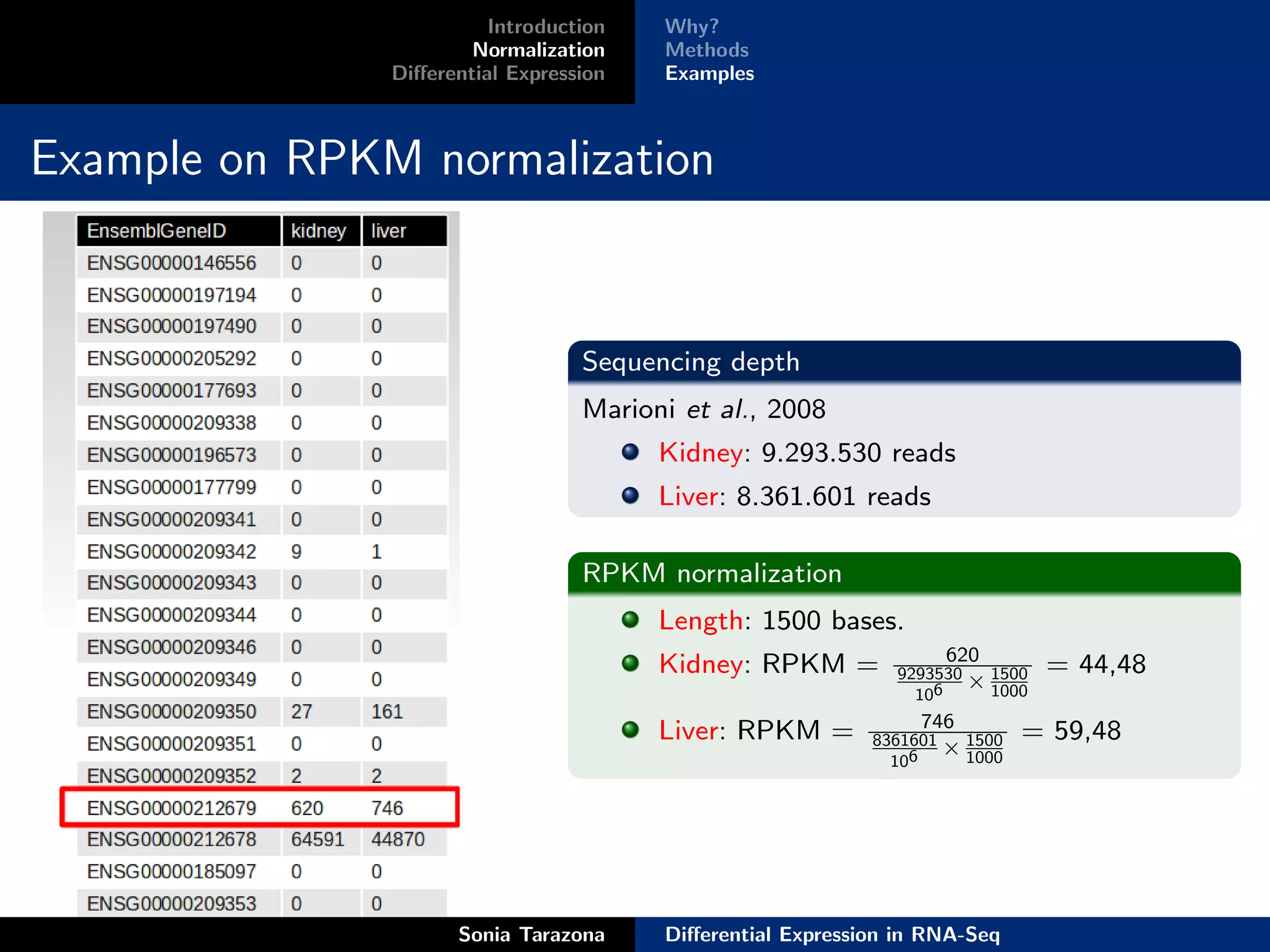
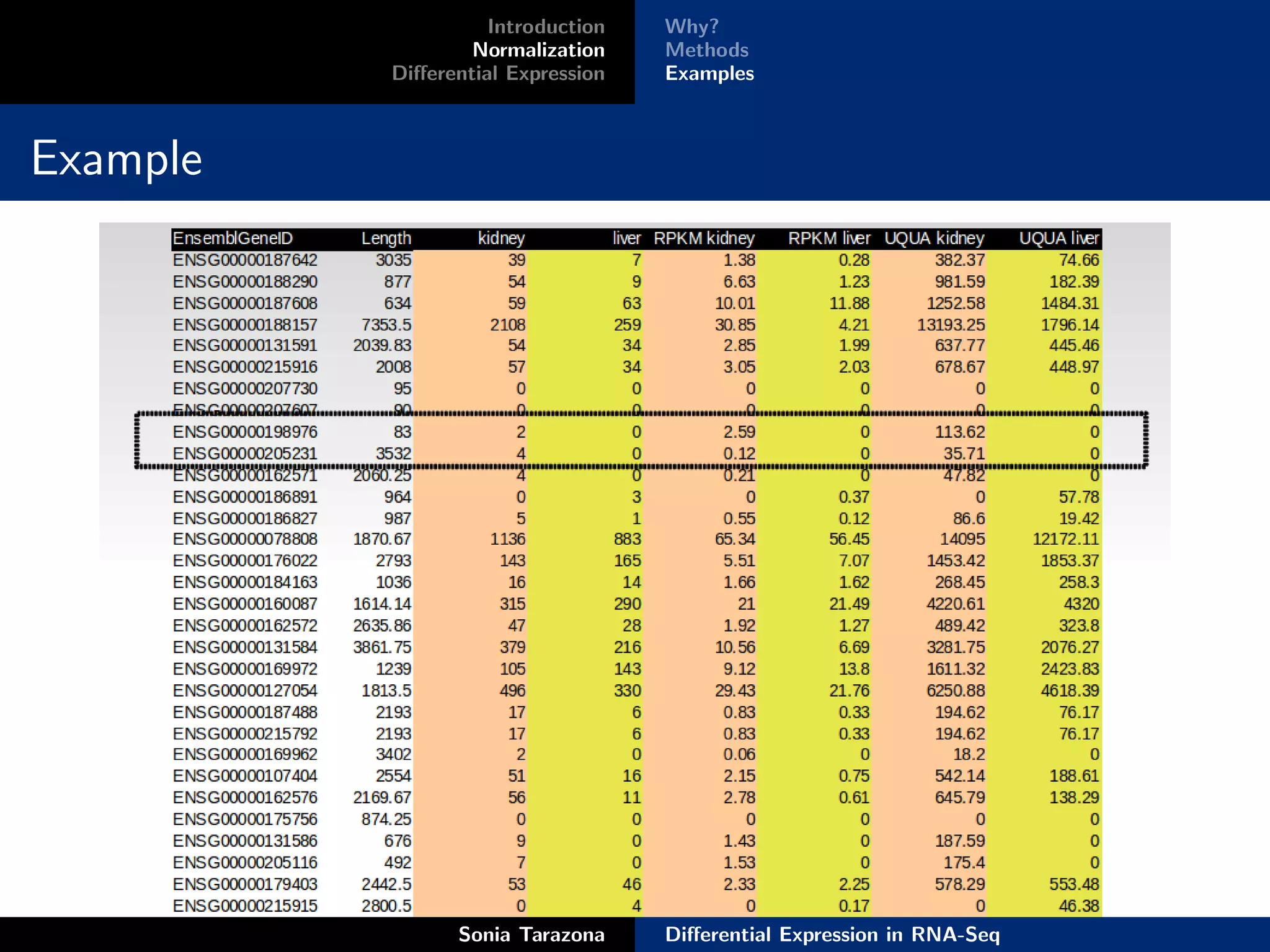
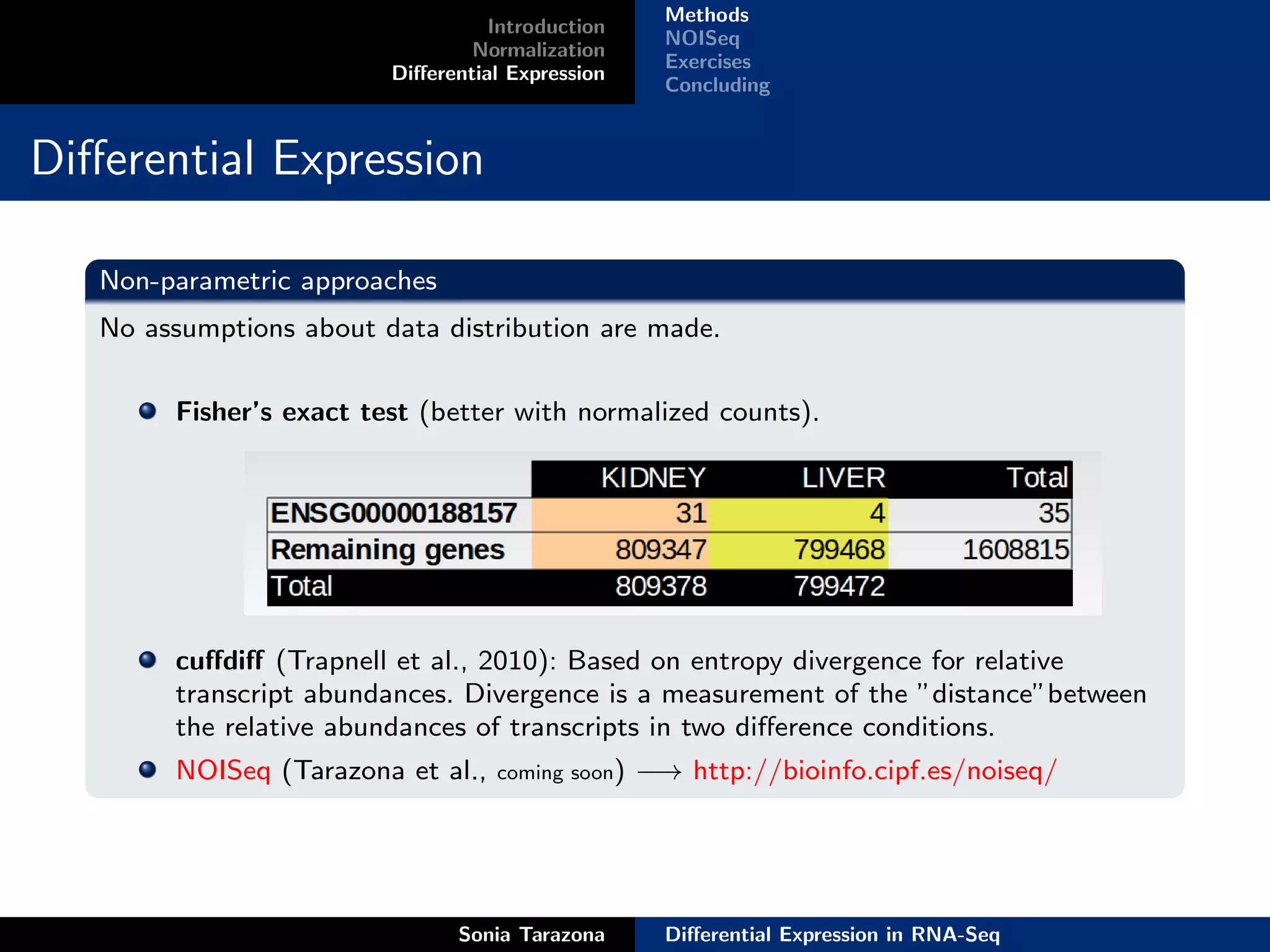
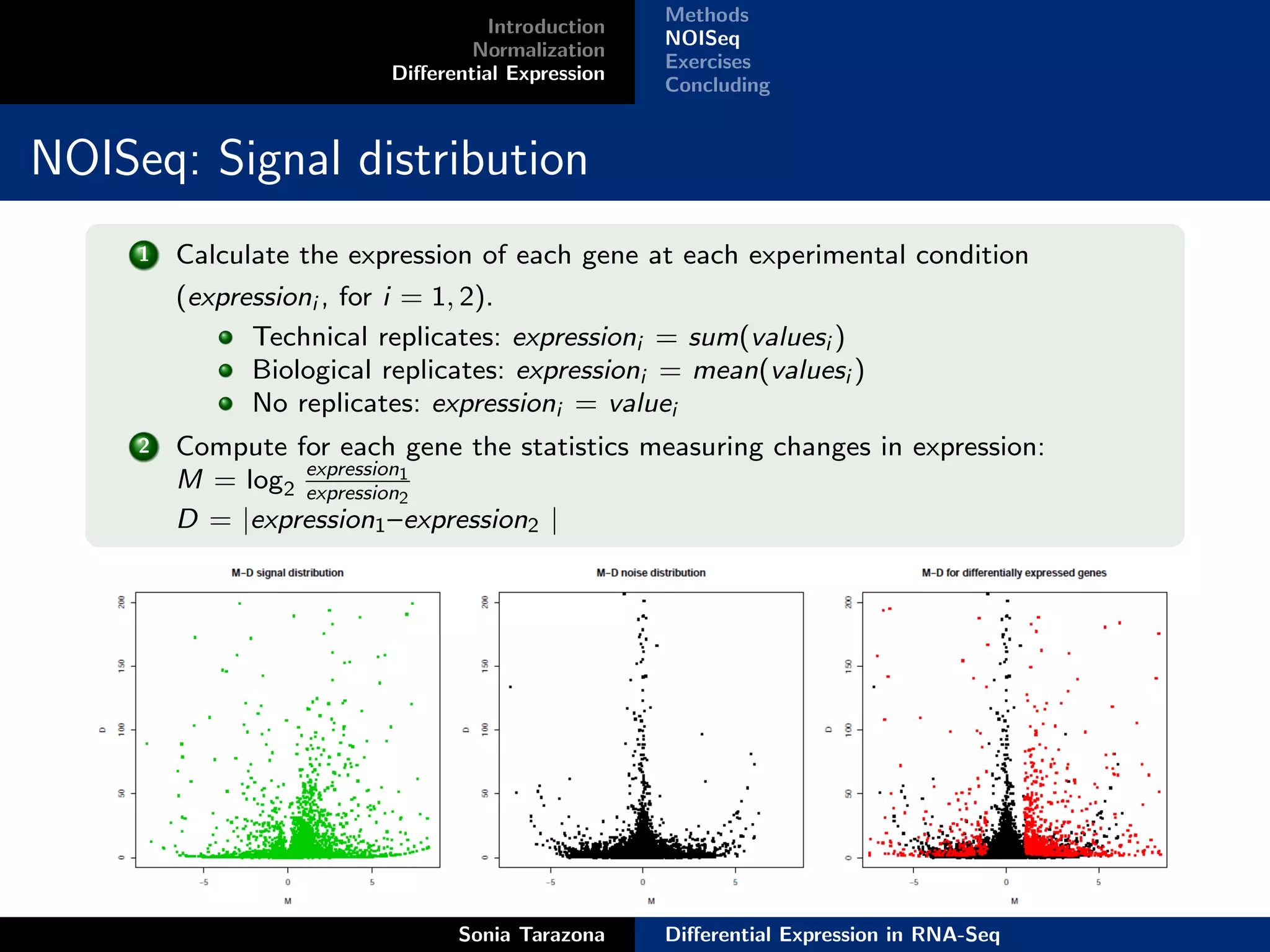
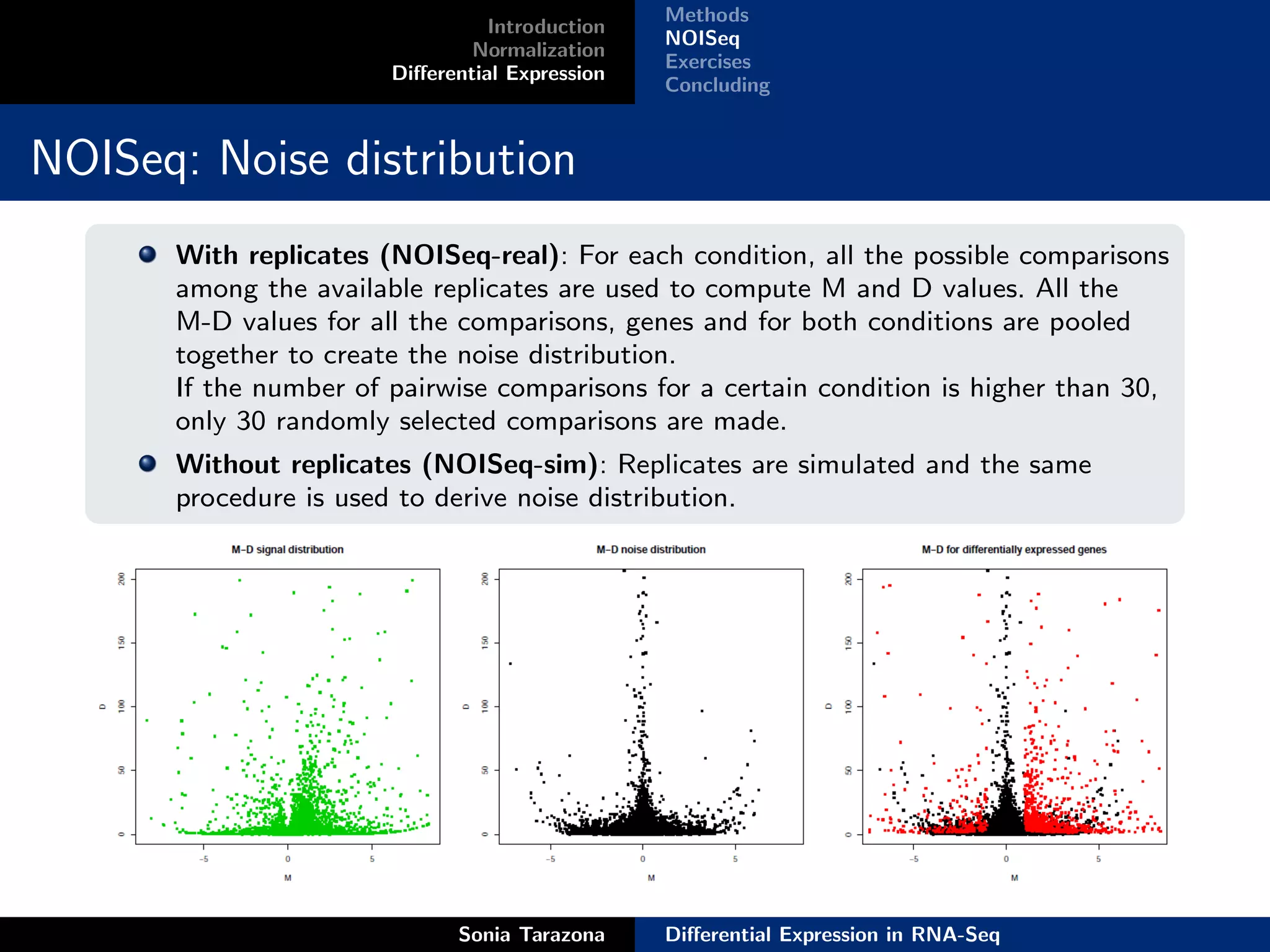
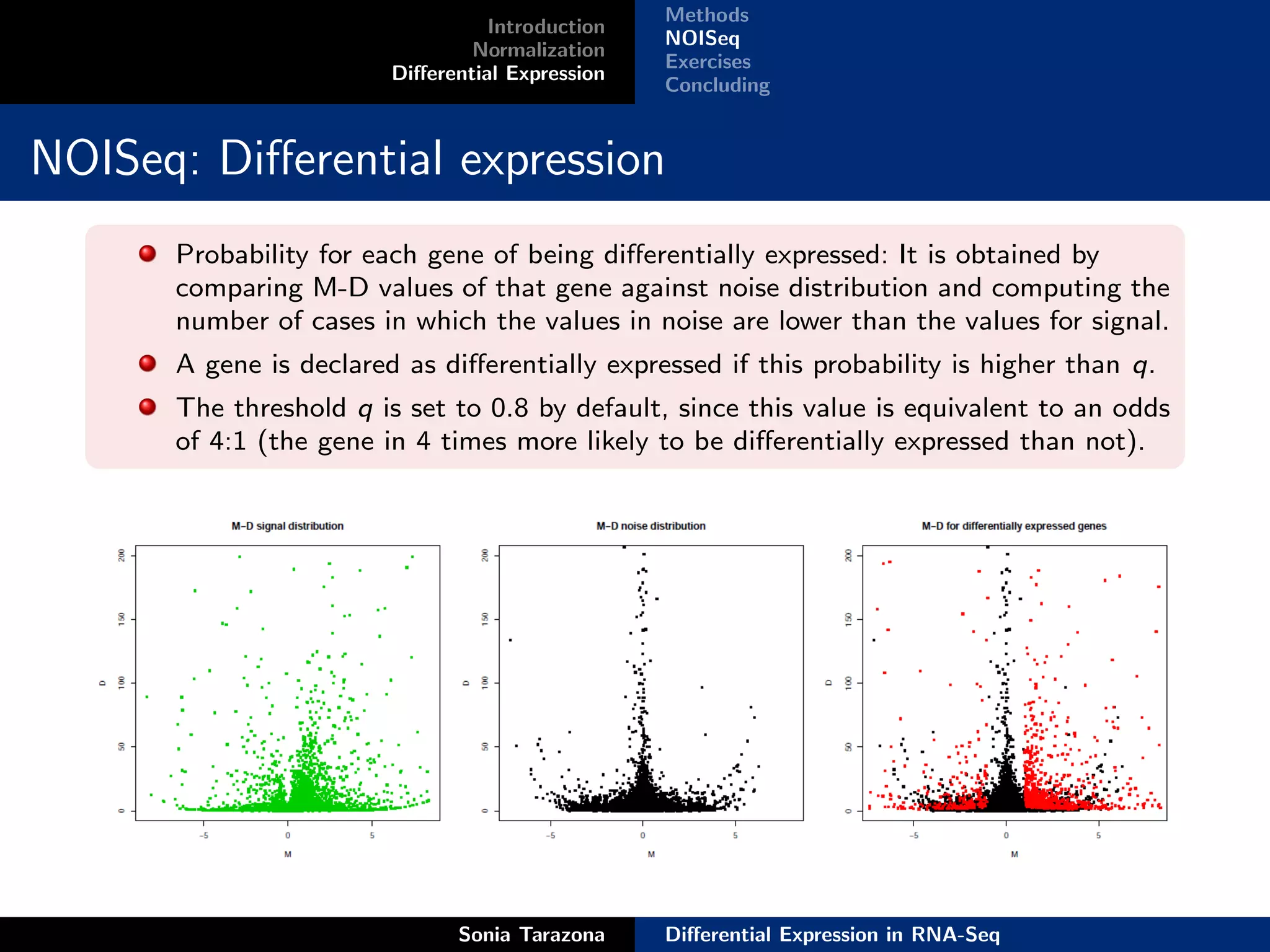
This document discusses differential expression analysis in RNA-Seq. It begins with an introduction that defines key concepts like expression levels, sequencing depth, and differential expression. It then covers normalization methods to account for biases in RNA-Seq data. The main method discussed is NOISeq, a non-parametric approach that does not require replicates. NOISeq compares signal distributions between conditions to noise distributions within conditions to identify differentially expressed genes. The document concludes with exercises to run NOISeq on sample data.








![Methods
Introduction
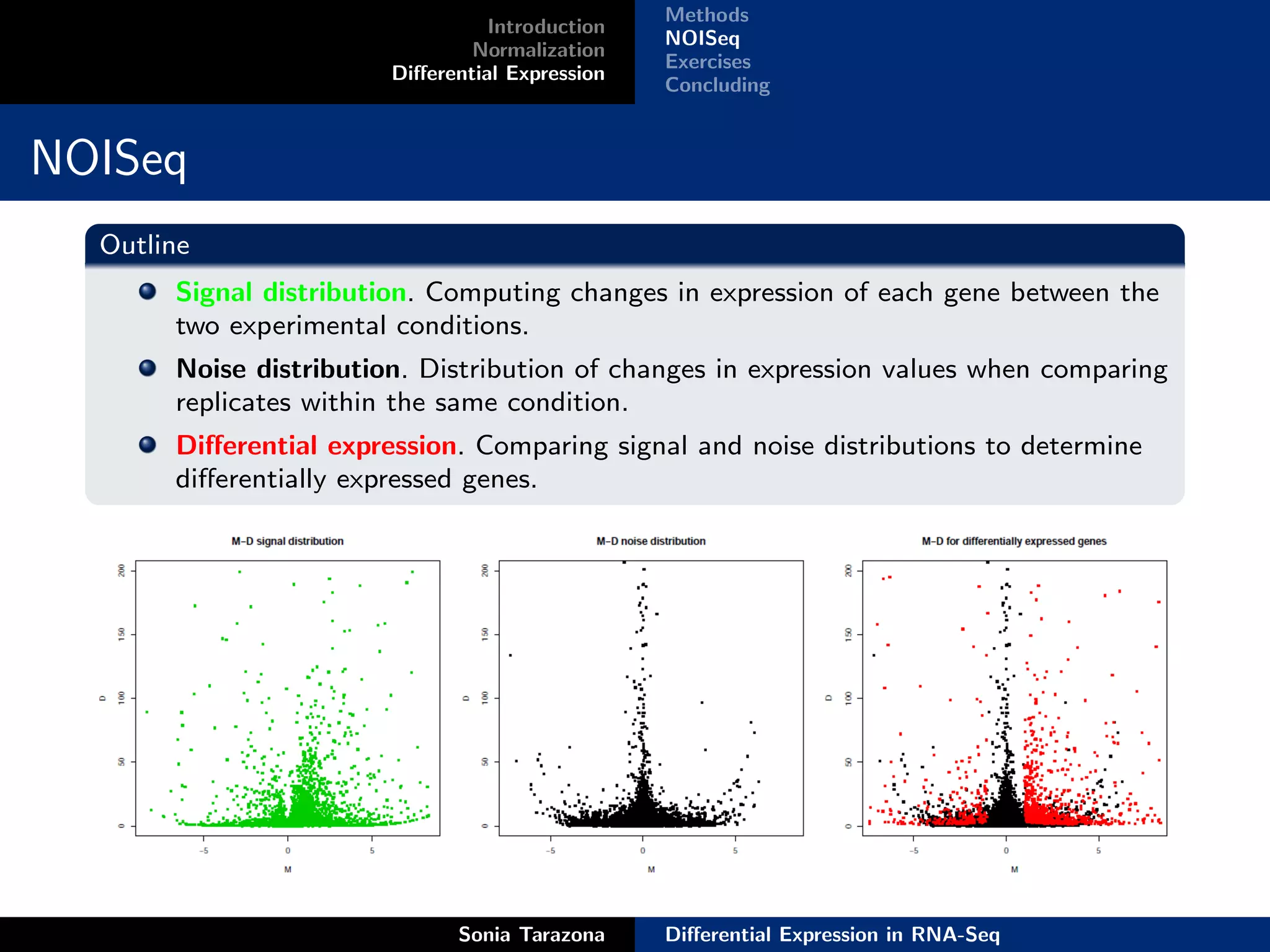
NOISeq
Normalization
Exercises
Differential Expression
Concluding
Exercises
Execute in an R terminal the code provided in exerciseDEngs.r
Reading data
simCount <- readData(file = "simCount.txt", cond1 = c(2:6),
cond2 = c(7:11), header = TRUE)
lapply(simCount, head)
depth <- as.numeric(sapply(simCount, colSums))
Differential Expression by NOISeq
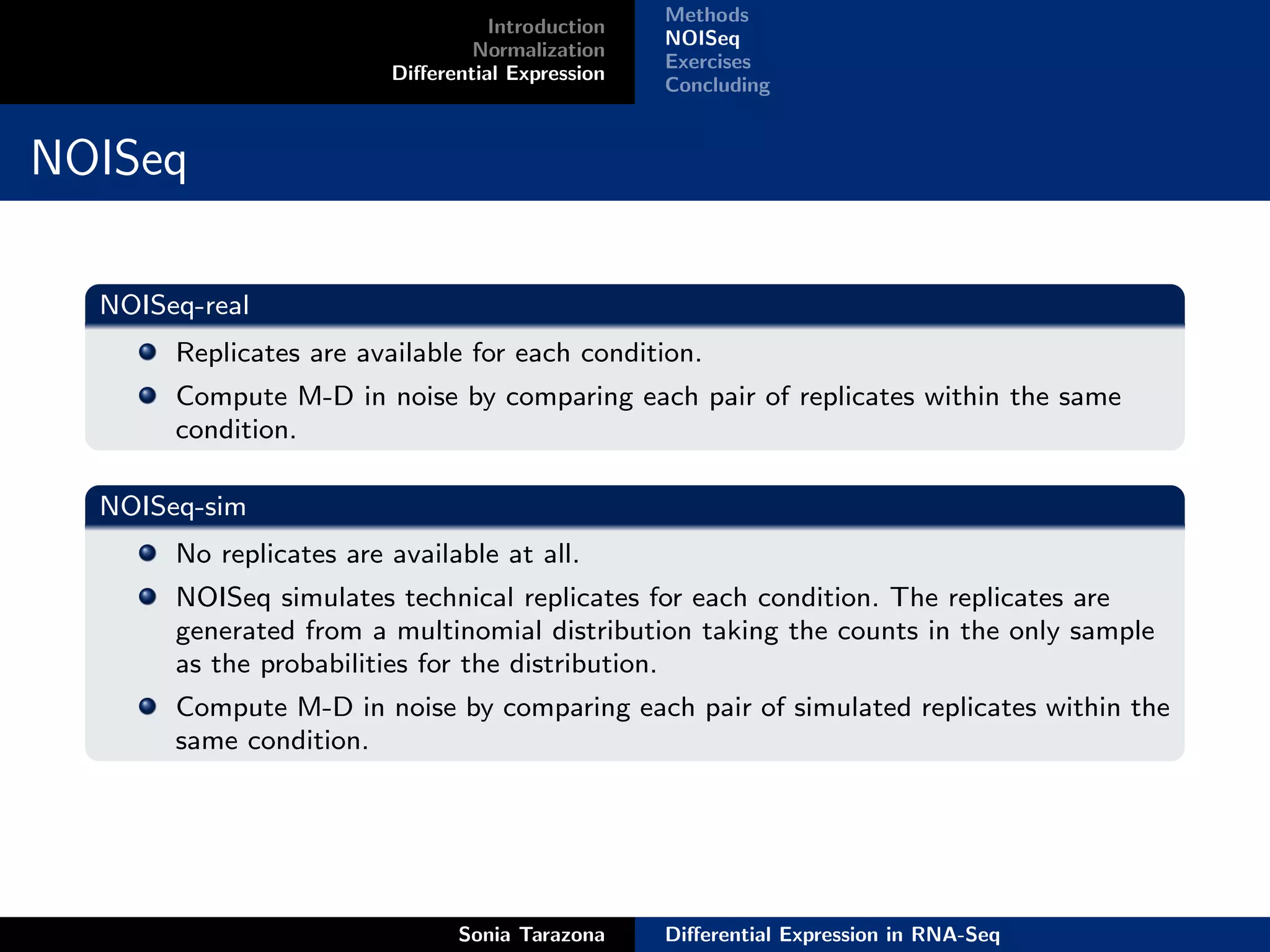
NOISeq-real
res1noiseq <- noiseq(simCount[[1]], simCount[[2]], nss = 0, q = 0.8,
repl = ‘‘tech’’)
NOISeq-sim
res2noiseq <- noiseq(as.matrix(rowSums(simCount[[1]])),
as.matrix(rowSums(simCount[[2]])), q = 0.8, pnr = 0.2, nss = 5)
See the tutorial in http://bioinfo.cipf.es/noiseq/ for more information.
Sonia Tarazona Differential Expression in RNA-Seq](https://image.slidesharecdn.com/soniatarazona-differentialexpression-110626051522-phpapp02/75/Differential-expression-in-RNA-Seq-30-2048.jpg)

