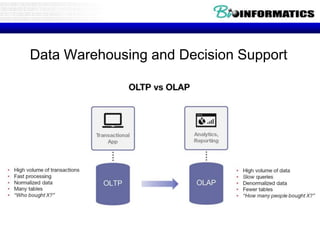
The document discusses views and materialized views in data warehousing and decision support systems. It covers three main points:
1) OLAP queries typically involve aggregate queries, so precomputation is essential for fast response times. Materialized views allow precomputing aggregates across multiple dimensions.
2) Warehouses can be thought of as collections of asynchronously replicated tables and periodically maintained views, renewing interest in efficient view maintenance.
3) Materialized views store the results of views in the database for fast access like a cache, but they require maintenance as underlying tables change. Incremental maintenance algorithms are ideal to efficiently update materialized views.




![View Materialization (Precomputation)
• Suppose we precompute RegionalSales and store
it with a clustered B+ tree index on
[category,state,sales].
– Then, previous query can be answered by an index-
only scan.
SELECT R.state, SUM(R.sales)
FROM RegionalSales R
WHERE R.category=“Laptop”
GROUP BY R.state
SELECT R.state, SUM(R.sales)
FROM RegionalSales R
WHERE R. state=“Wisconsin”
GROUP BY R.category
Index on precomputed view
is great!
Index is less useful (must
scan entire leaf level).](https://image.slidesharecdn.com/20190305biologicaldatabasespart4vupload-190312221240/85/2019-03-05_biological_databases_part4_v_upload-7-320.jpg)

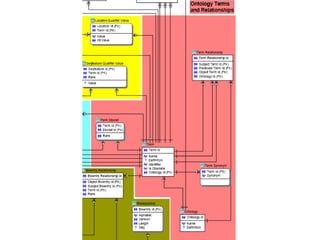







![Import from BioPython to BIOSQL
#Connecting to a BioSQL database -http://biopython.org/wiki/BioSQL
from Bio import Entrez
from Bio import SeqIO
from BioSQL import BioSeqDatabase
server = BioSeqDatabase.open_database(driver = "pymysql",host =
"localhost",port=8889,user="root",passwd="root",db="bio2019")
db = server.new_database("test2")
db = server["test2"]
import pprint
Entrez.email = "A.N.Other@example.com"
handle = Entrez.efetch(db="nucleotide", rettype="gb", retmode="text", id="6273291,6273290,6273289")
print ("Loading into BIOSQL")
count = db.load(SeqIO.parse(handle, "genbank"))
print ("Loaded %i records" % count)
server.adaptor.commit()
print ("ended succesfully")](https://image.slidesharecdn.com/20190305biologicaldatabasespart4vupload-190312221240/85/2019-03-05_biological_databases_part4_v_upload-25-320.jpg)


























![Lab for Bioinformatics and computational genomics
Overview
• Who ? Where ?
• > Genetics
• Technology: Next Gen Sequencing
• Personal …. Medicine/Genomics
• Manifesto
• The App
^[now][transl⎮comput]ational[epi]genomic$](https://image.slidesharecdn.com/20190305biologicaldatabasespart4vupload-190312221240/85/2019-03-05_biological_databases_part4_v_upload-57-320.jpg)