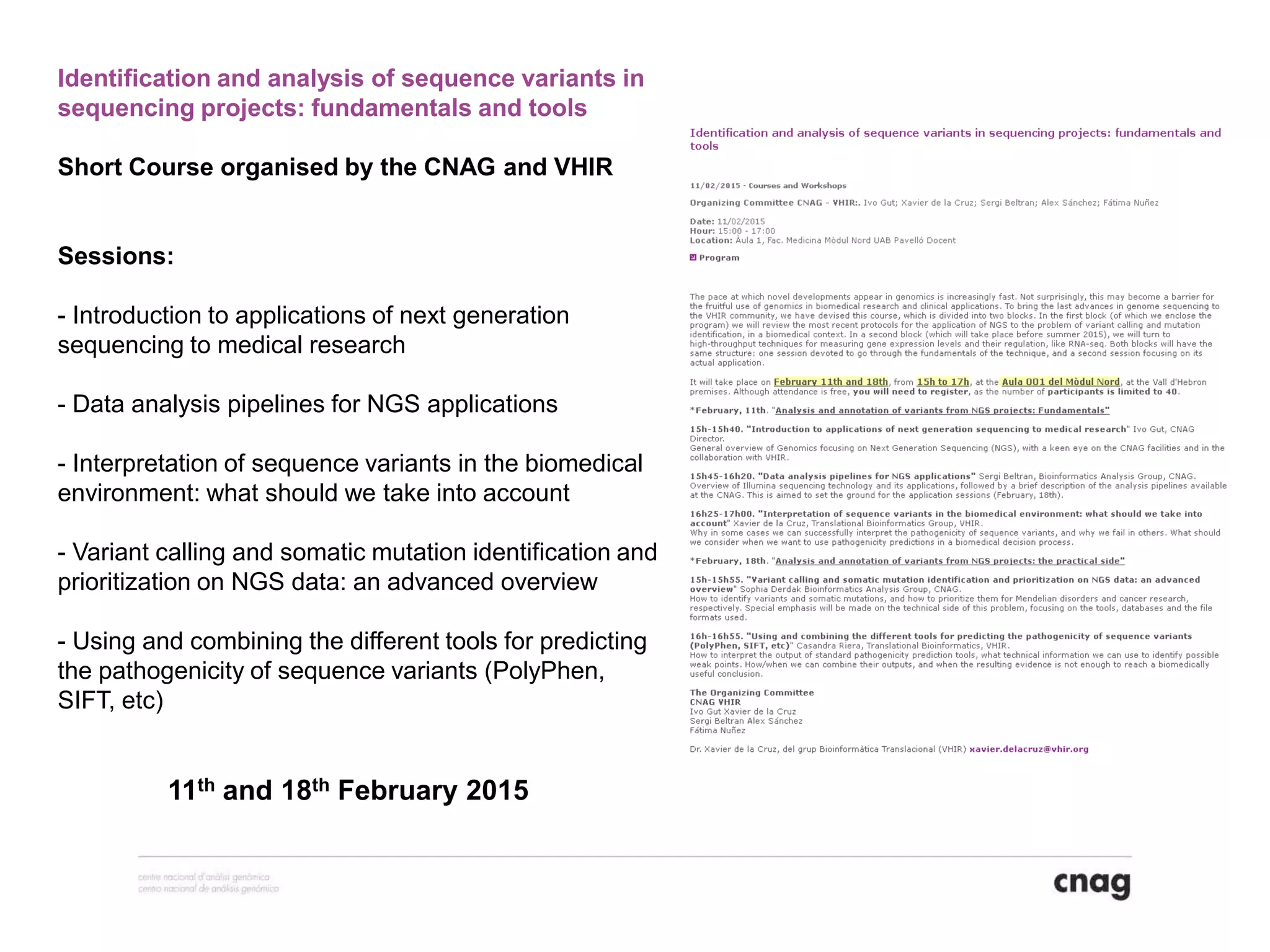
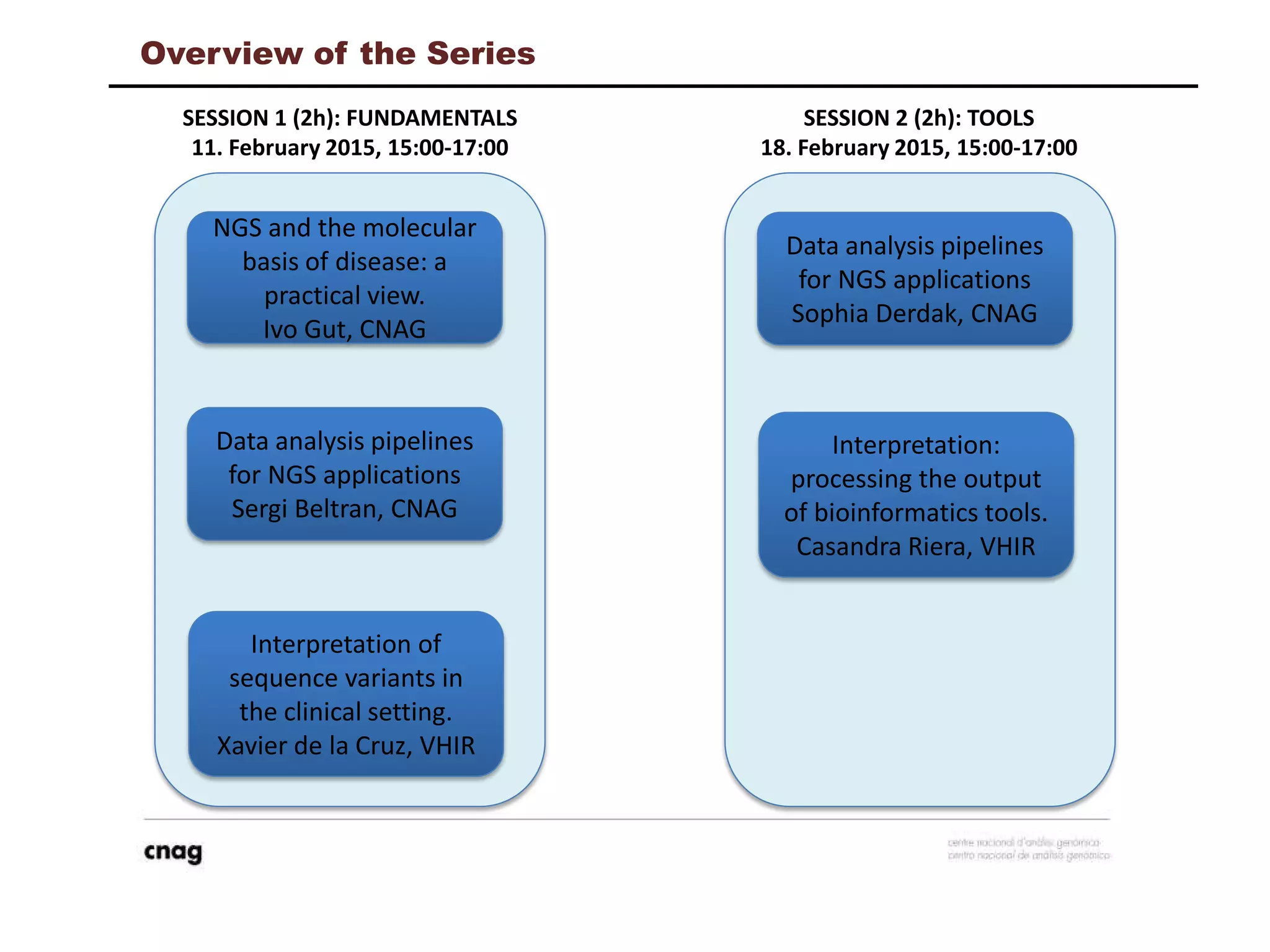
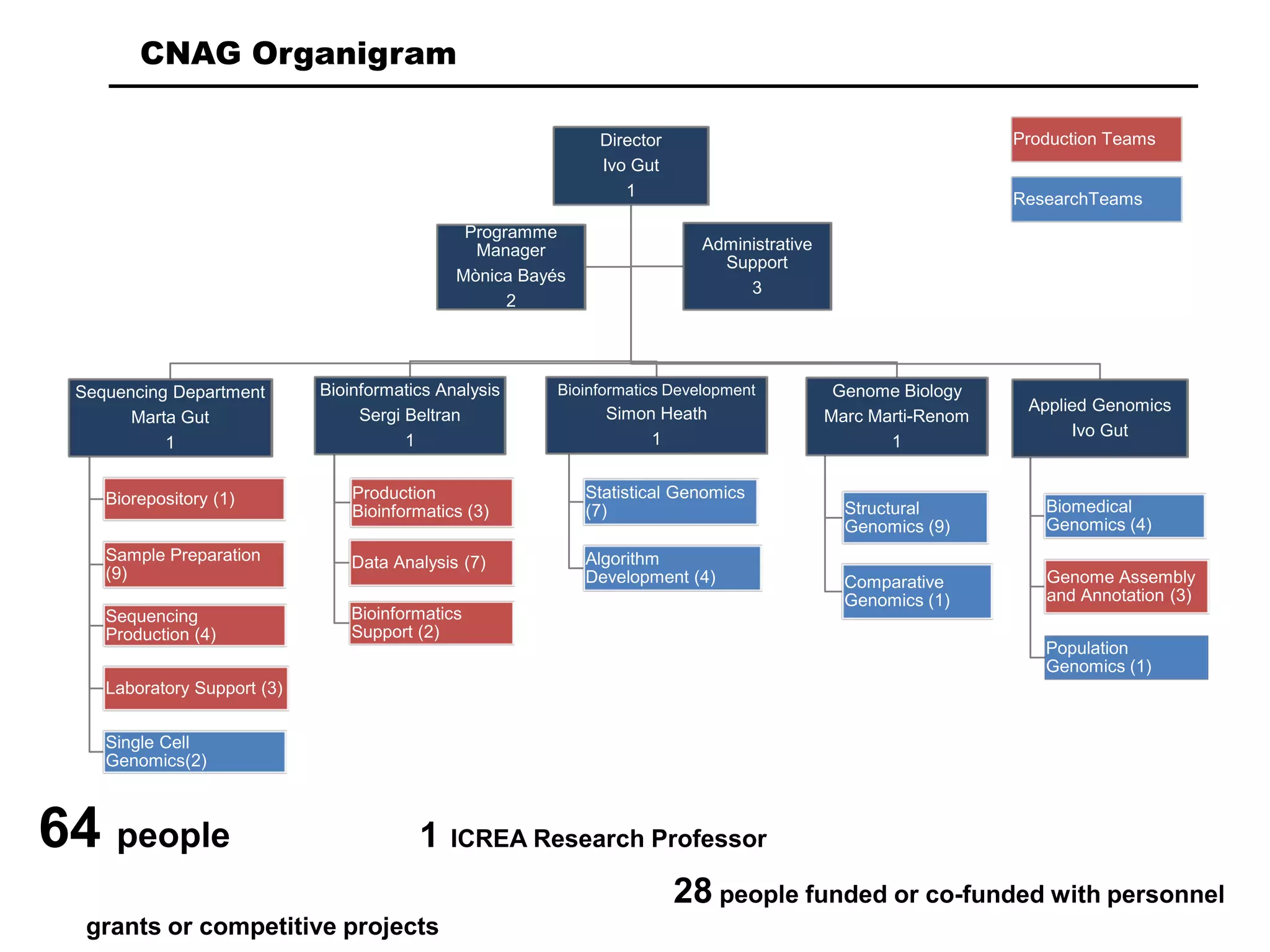
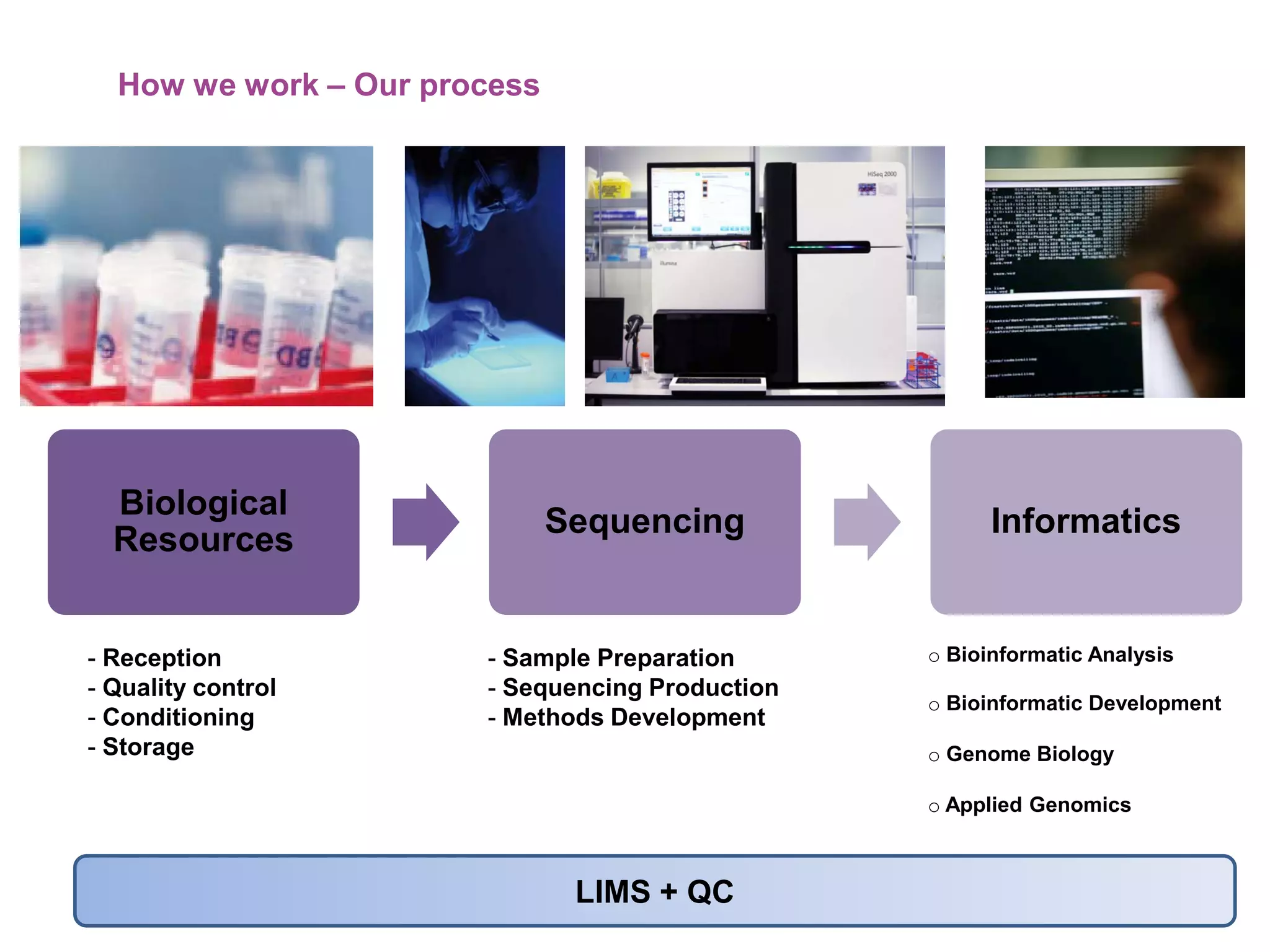
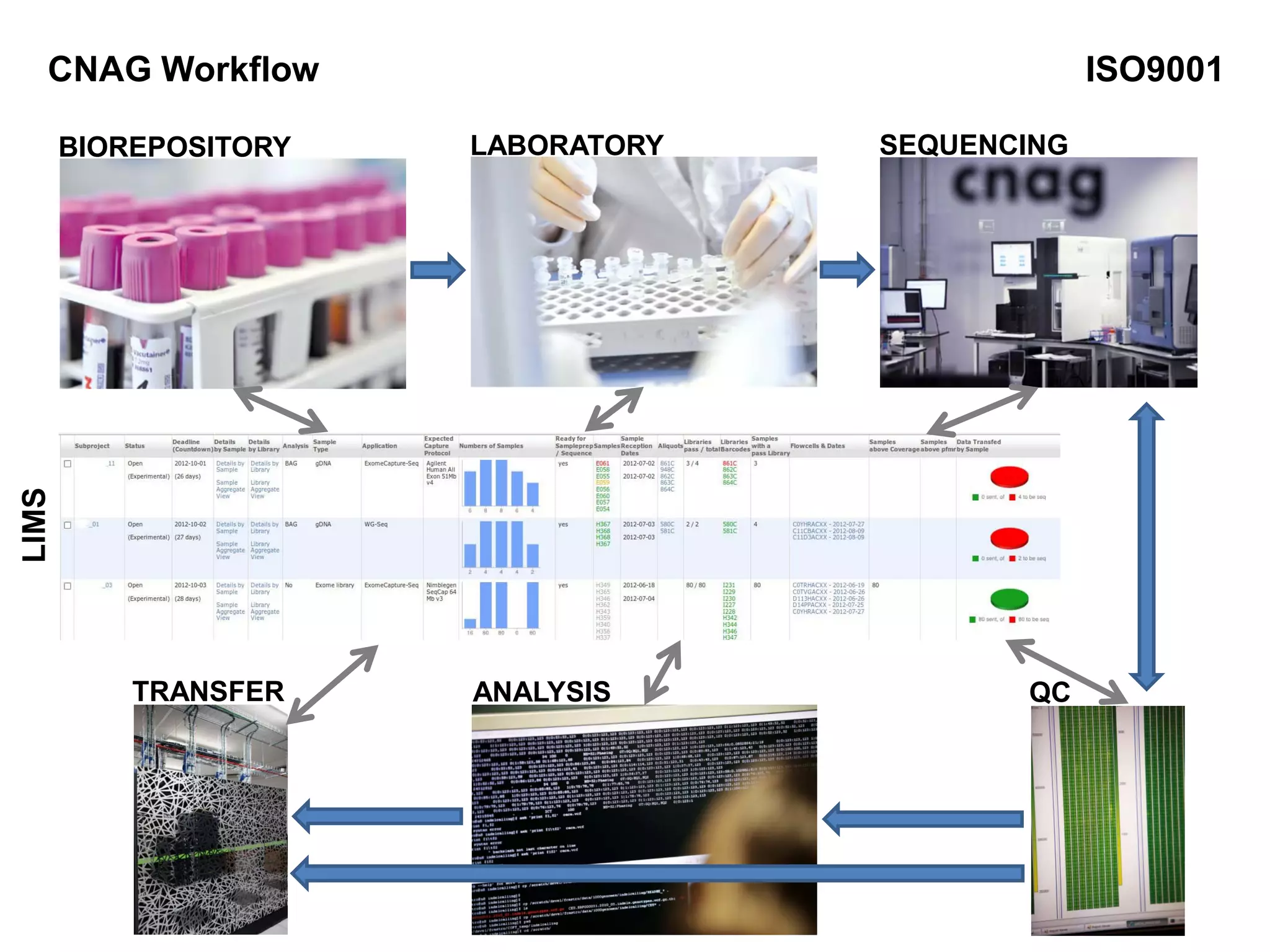
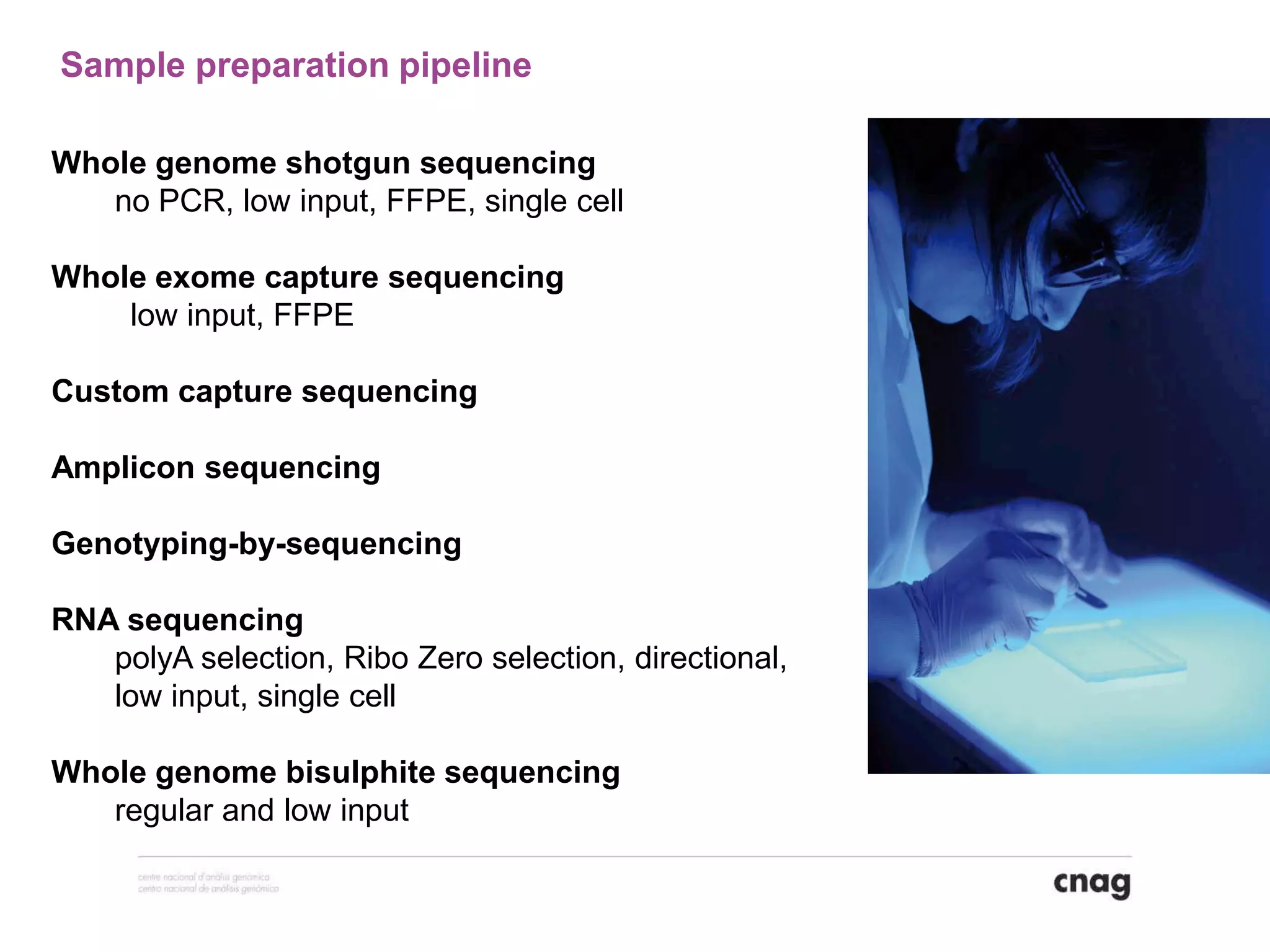
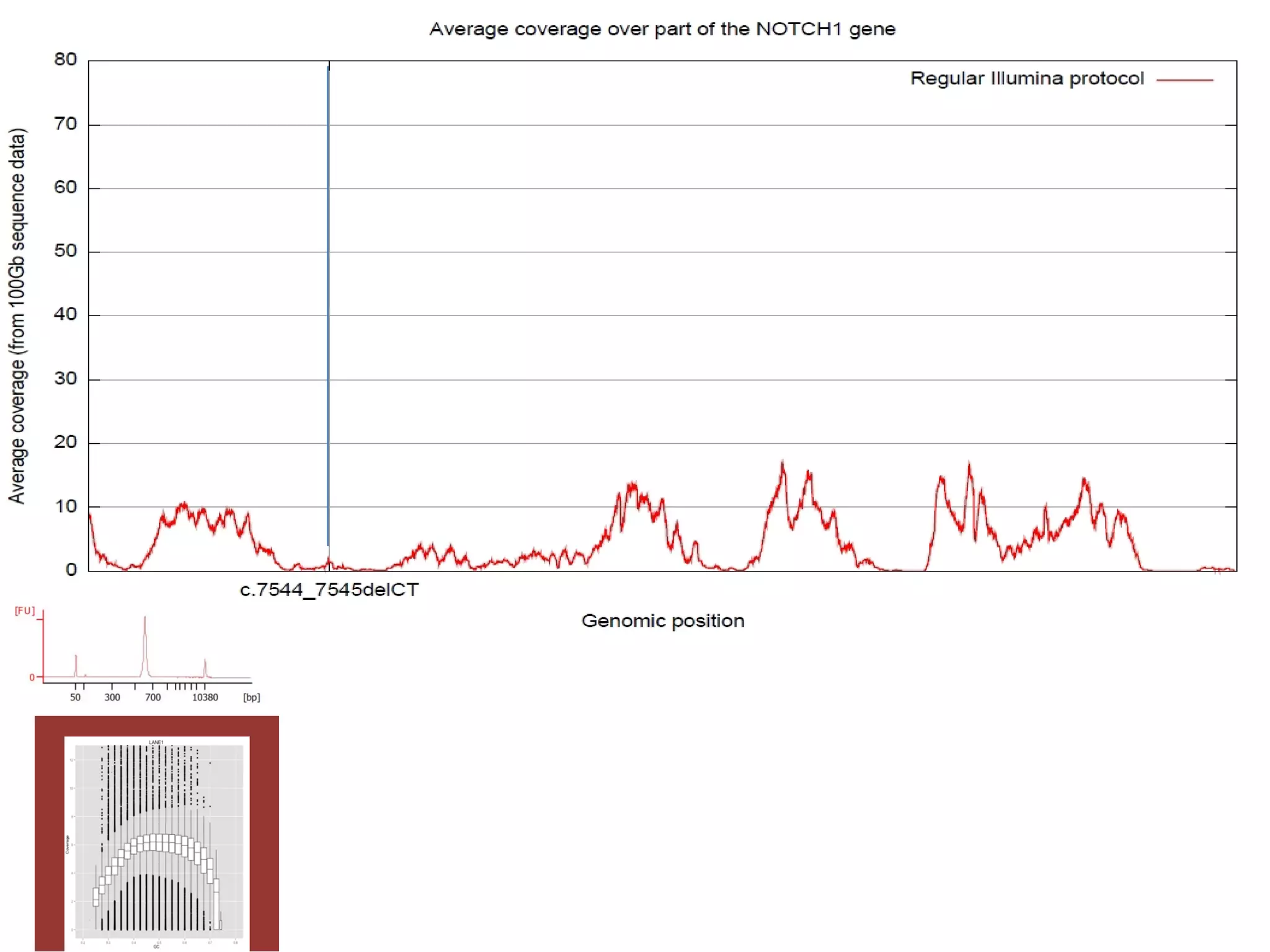
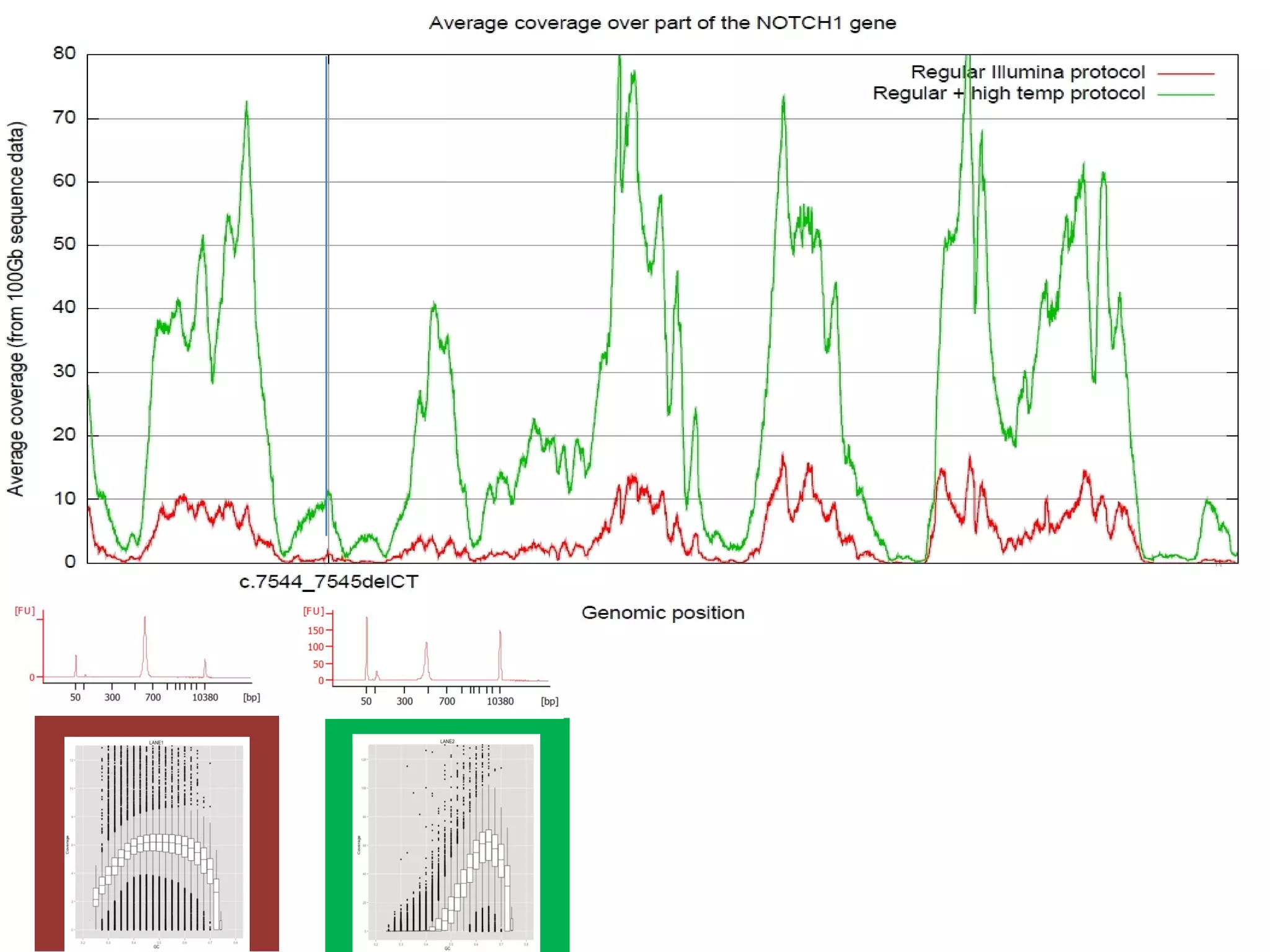
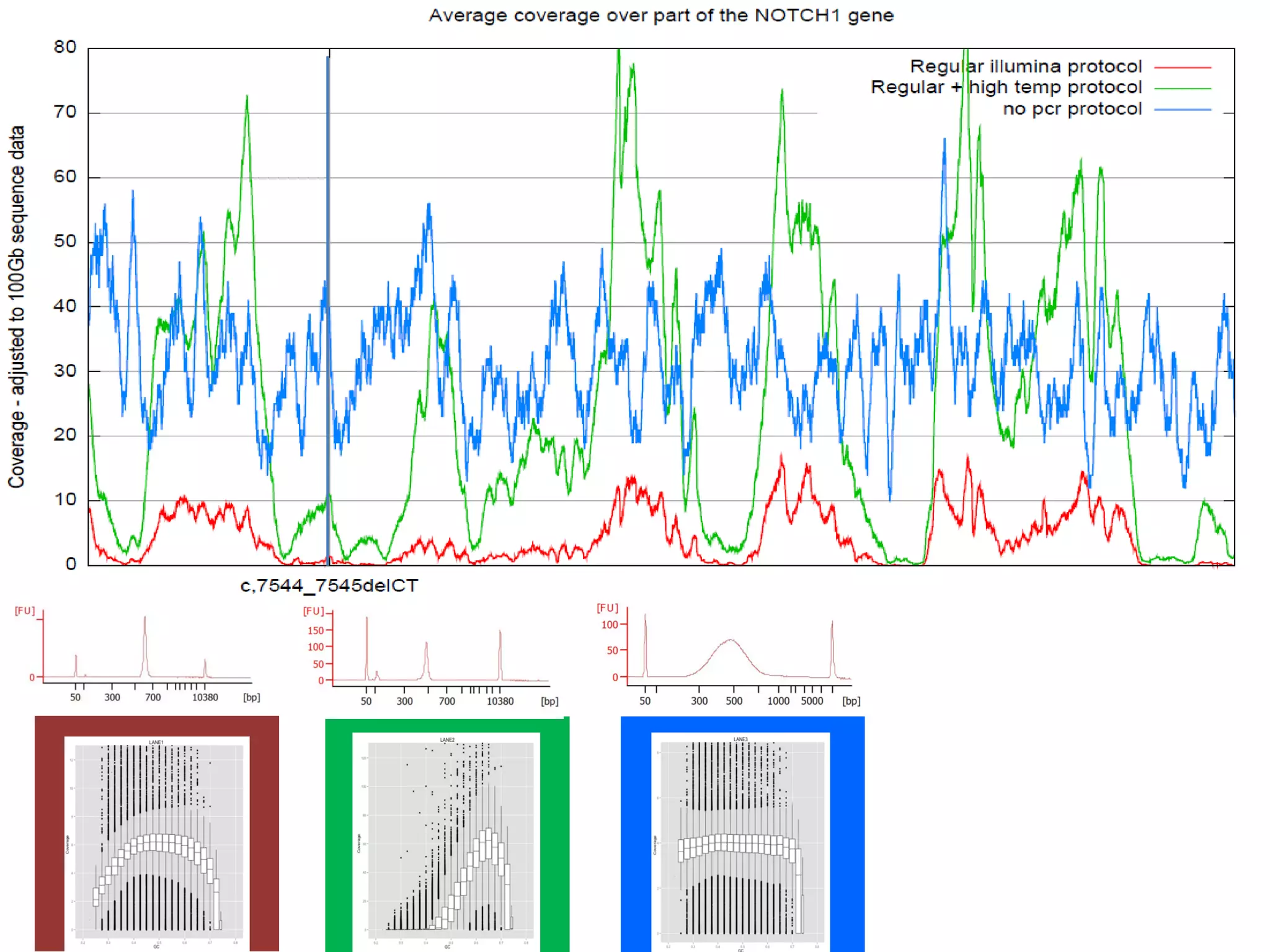
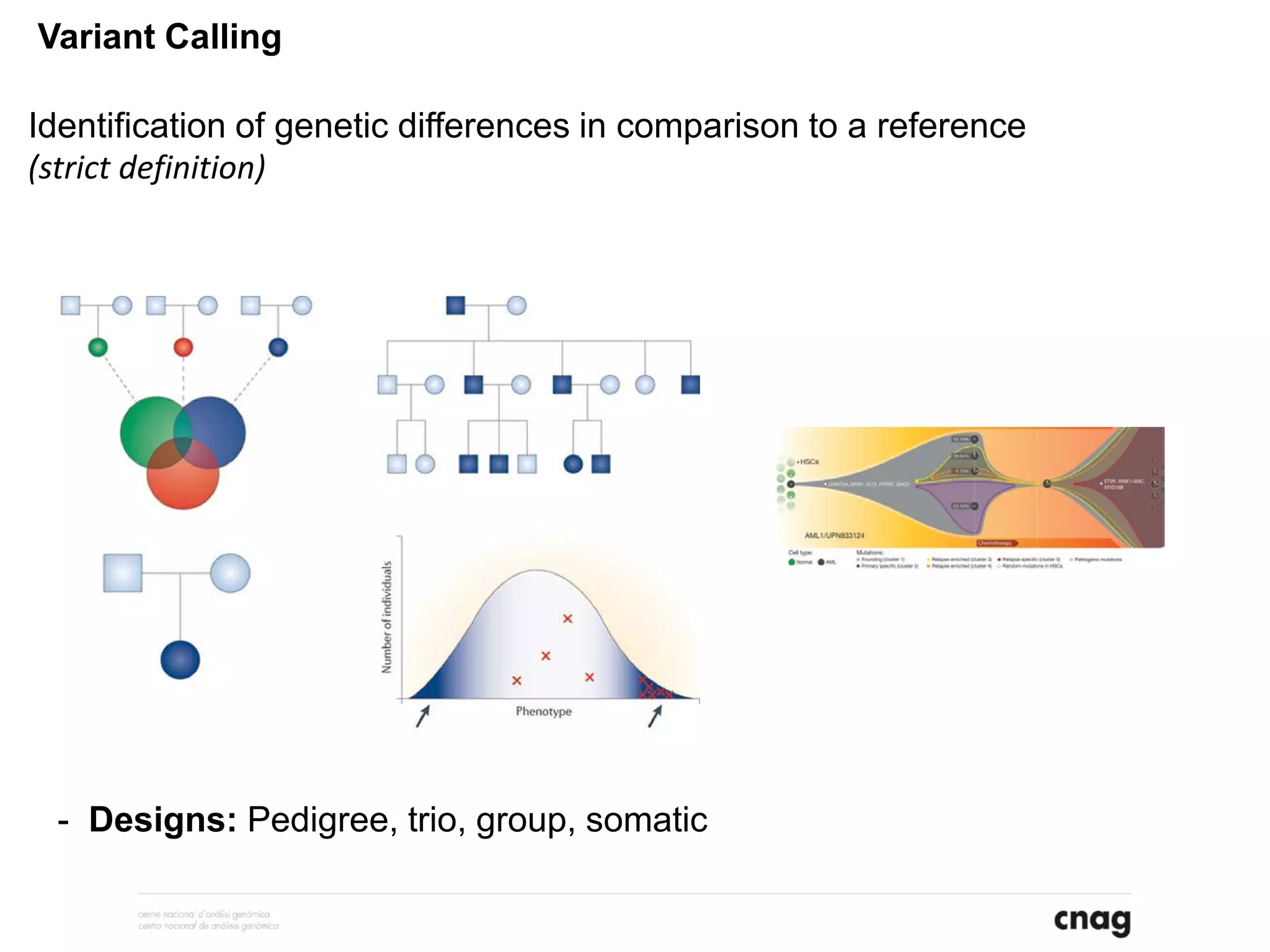
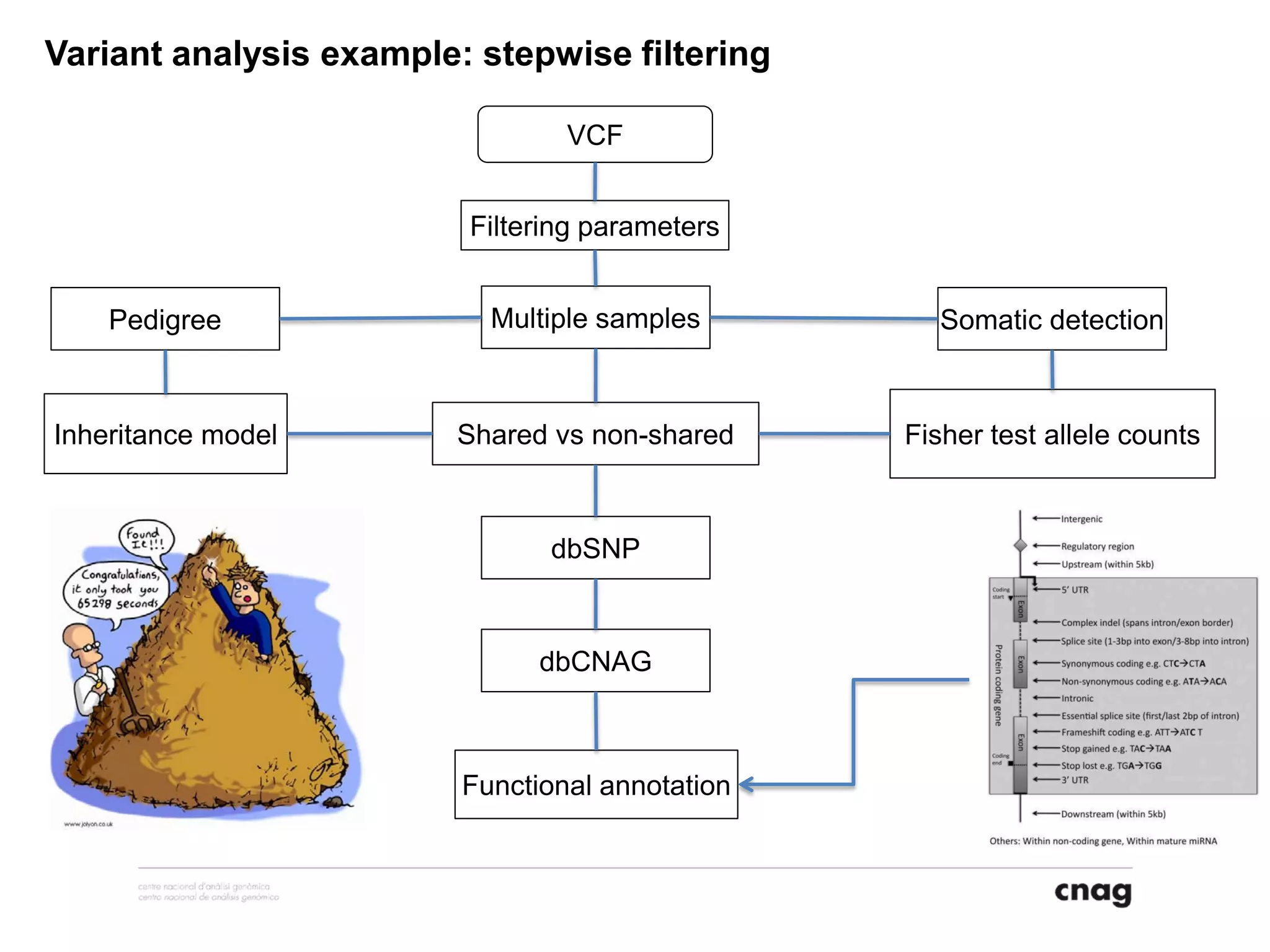
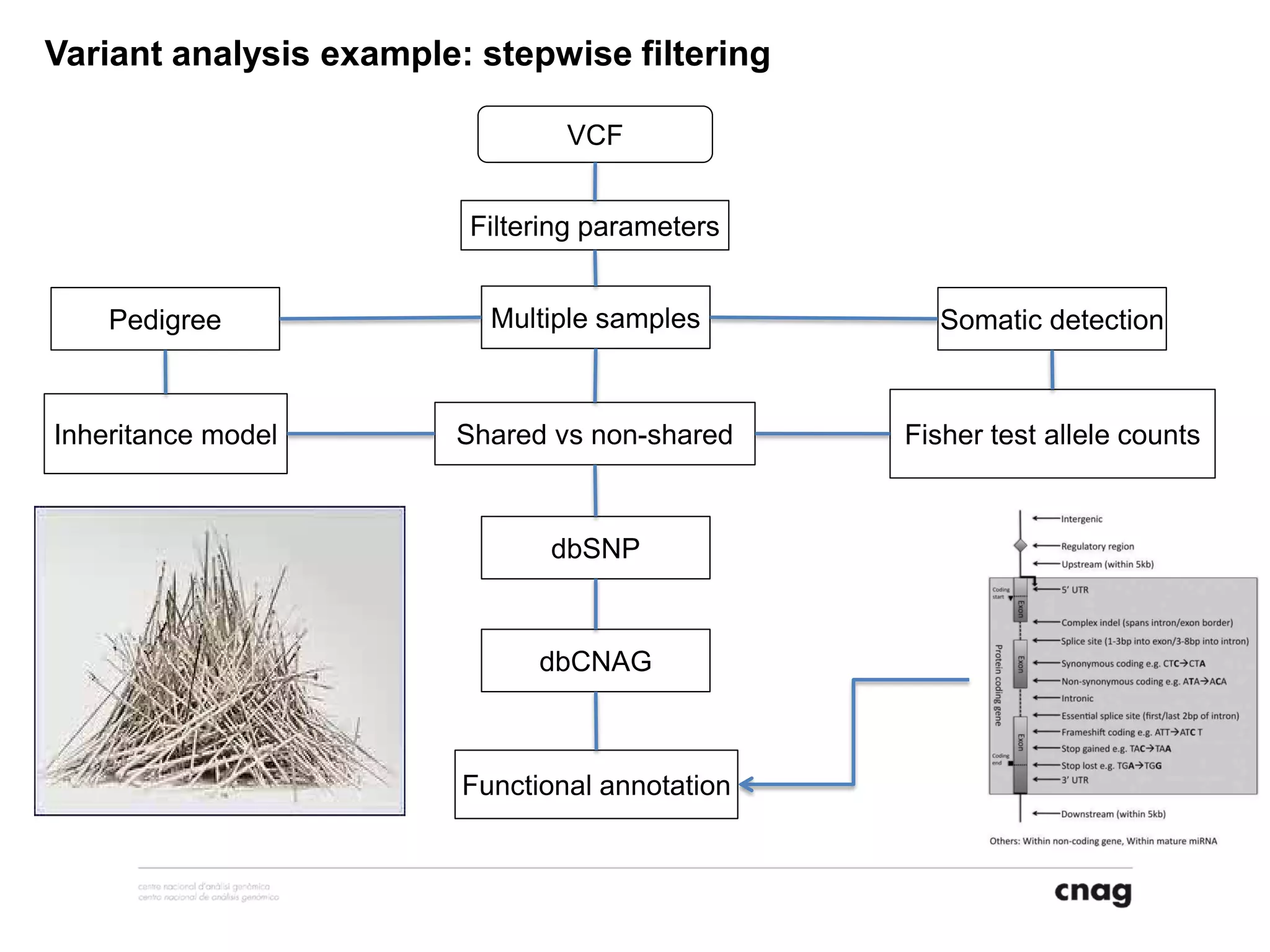
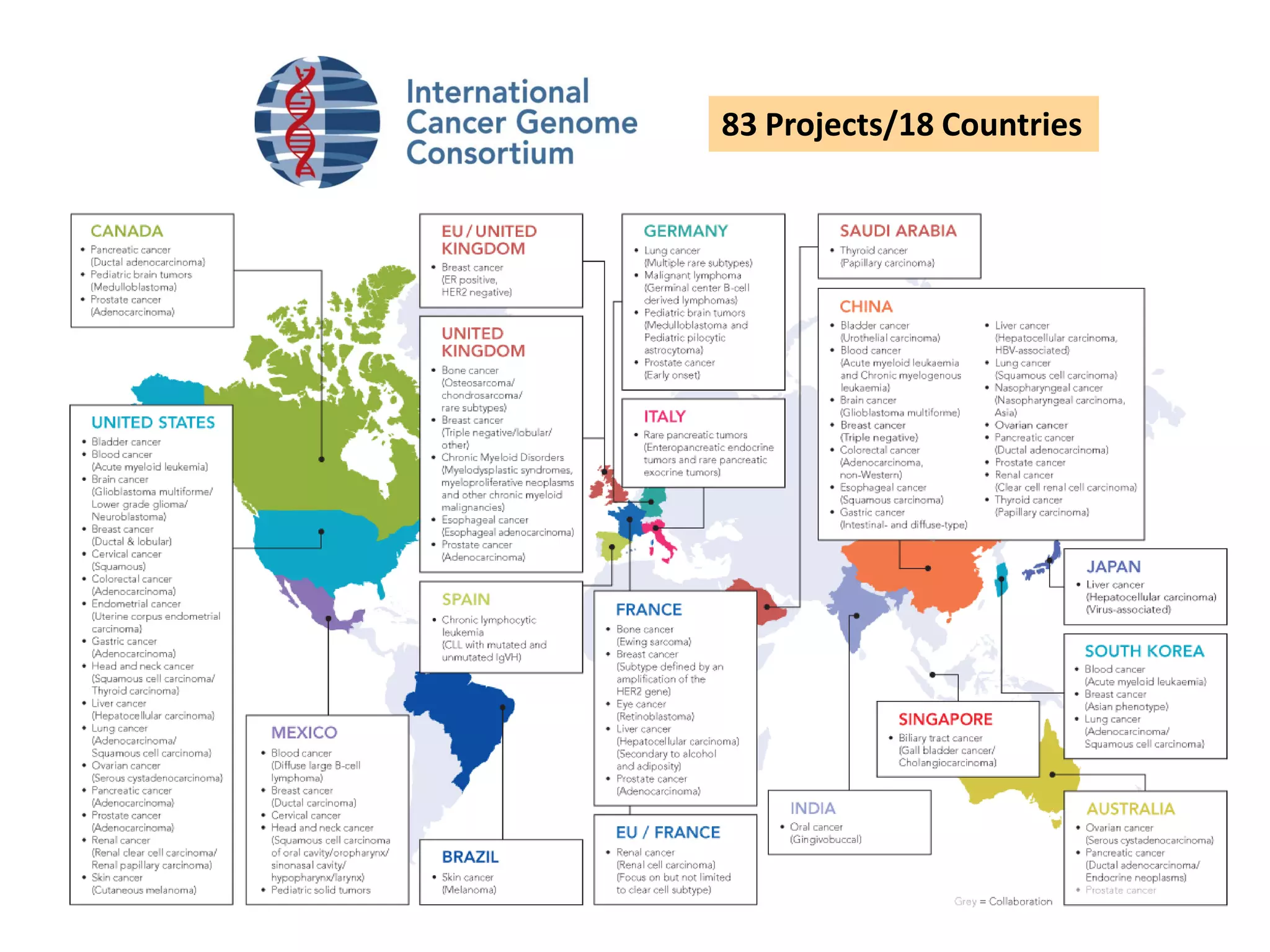
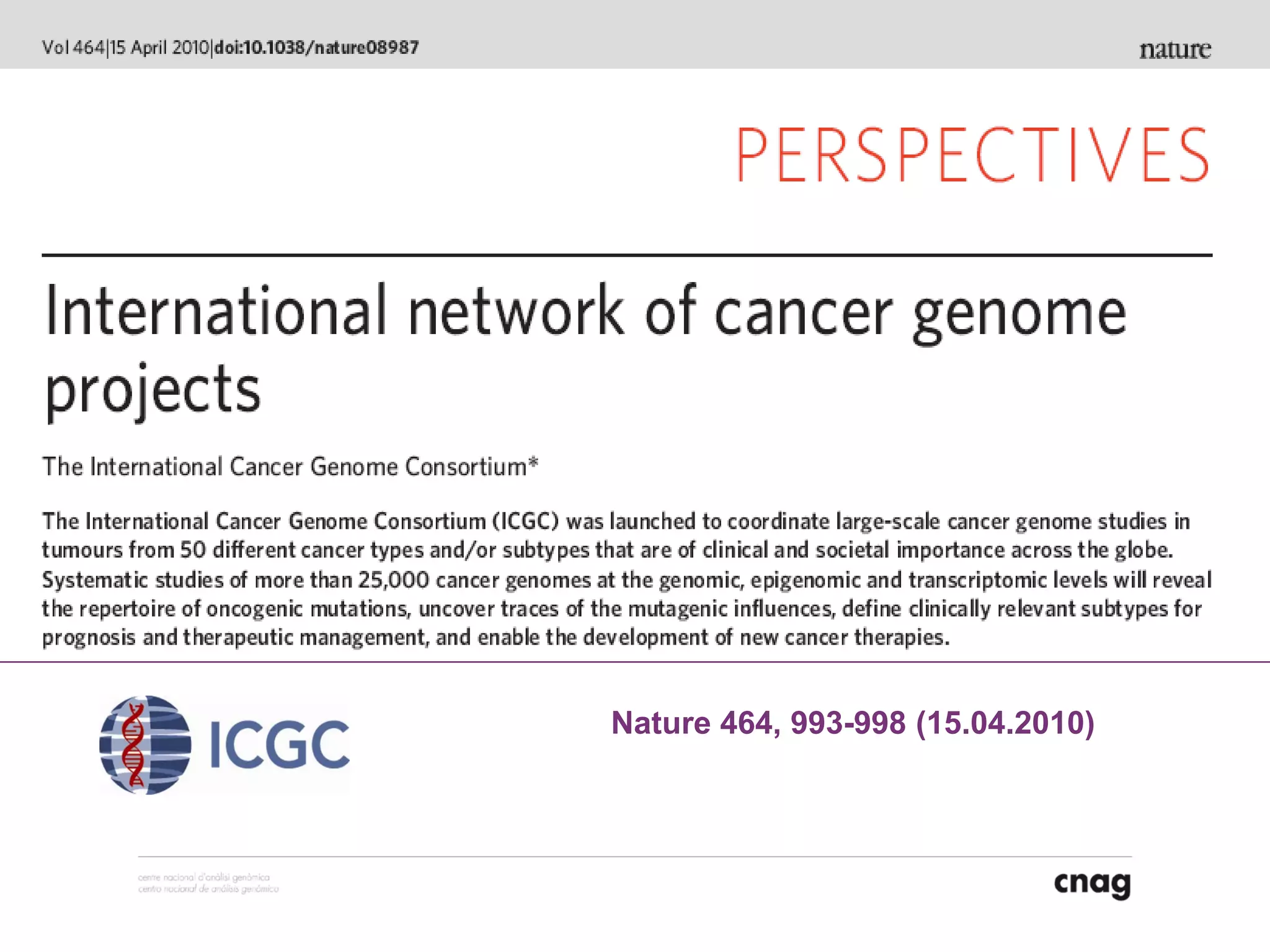
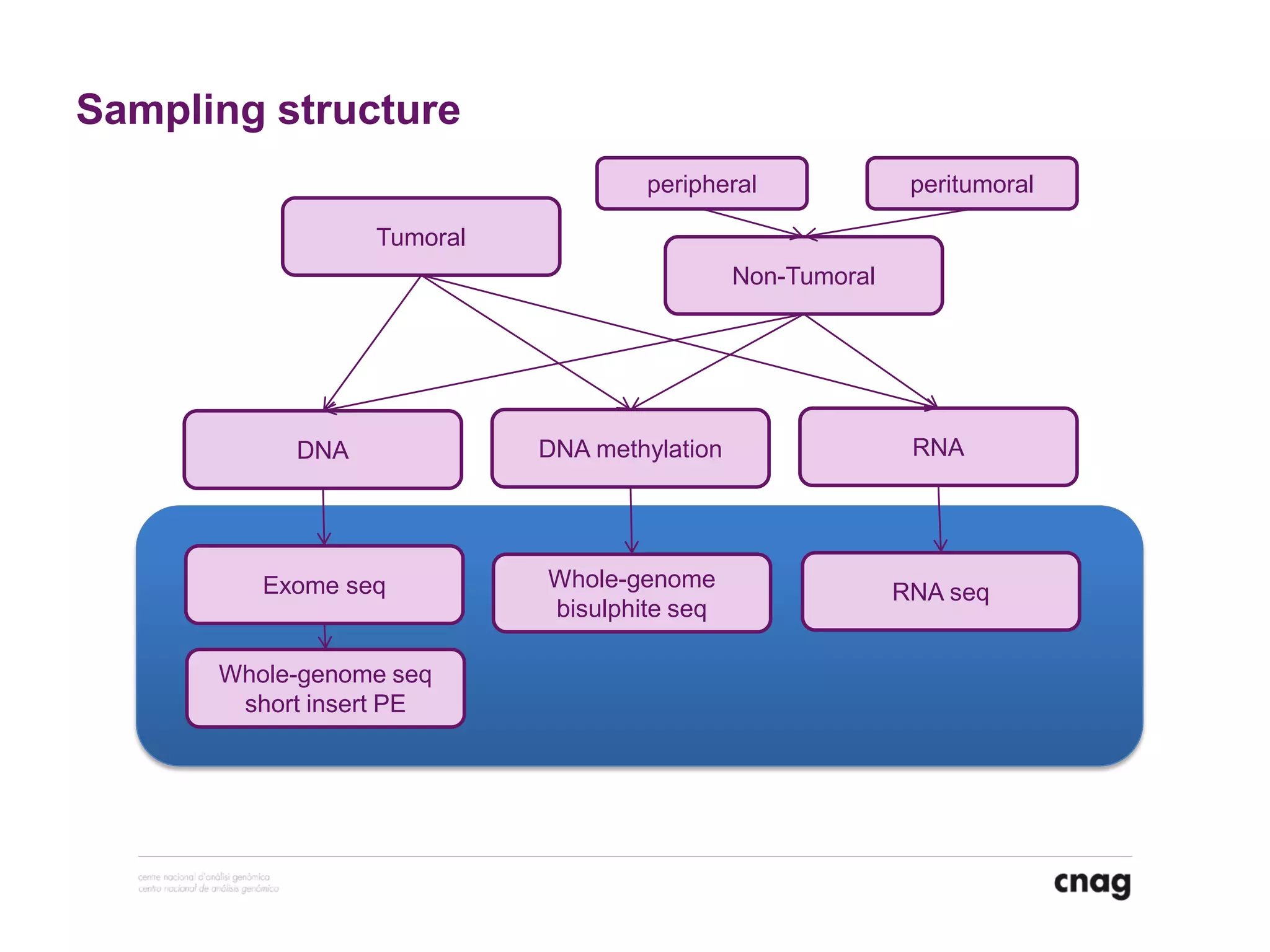
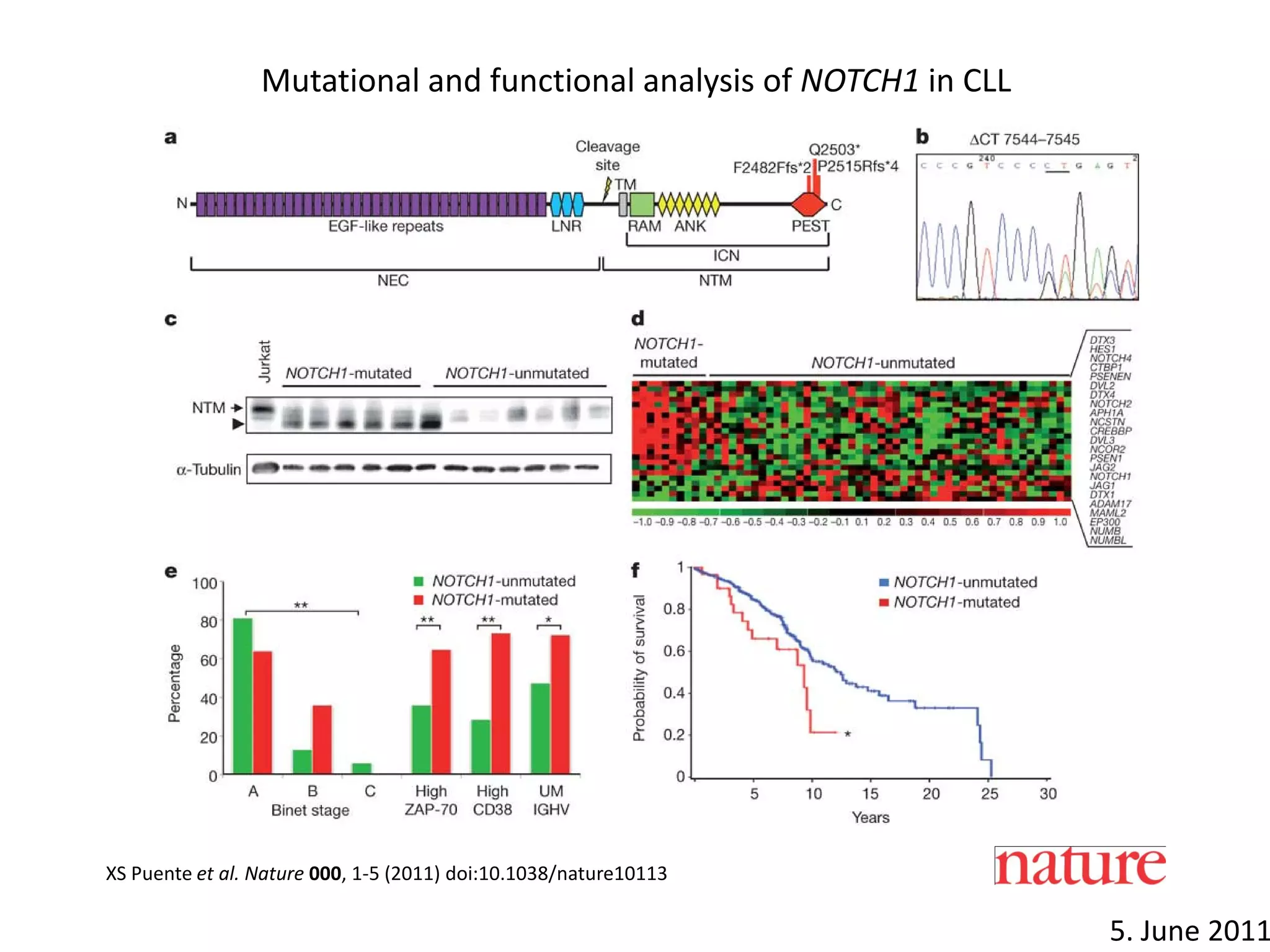
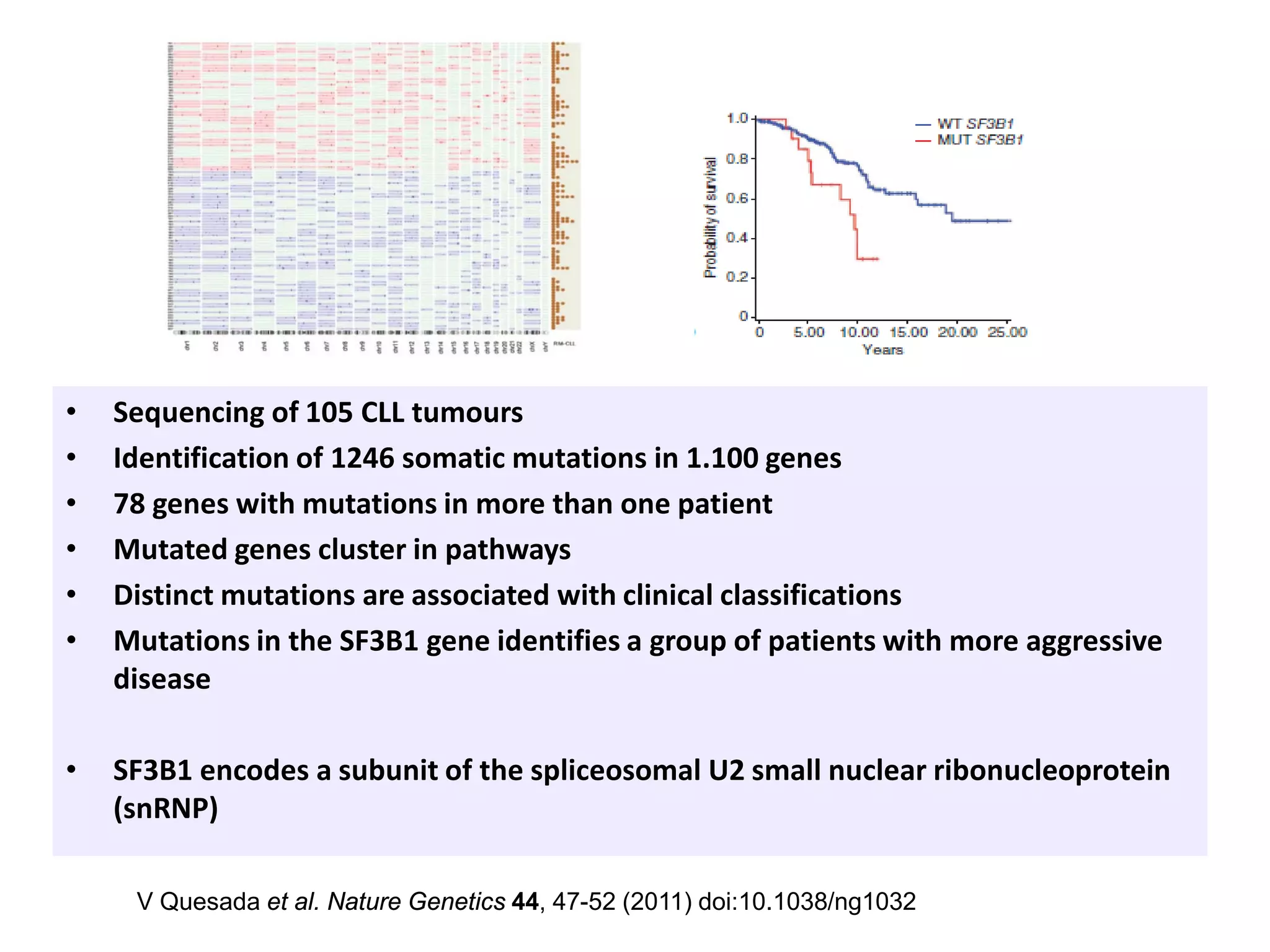
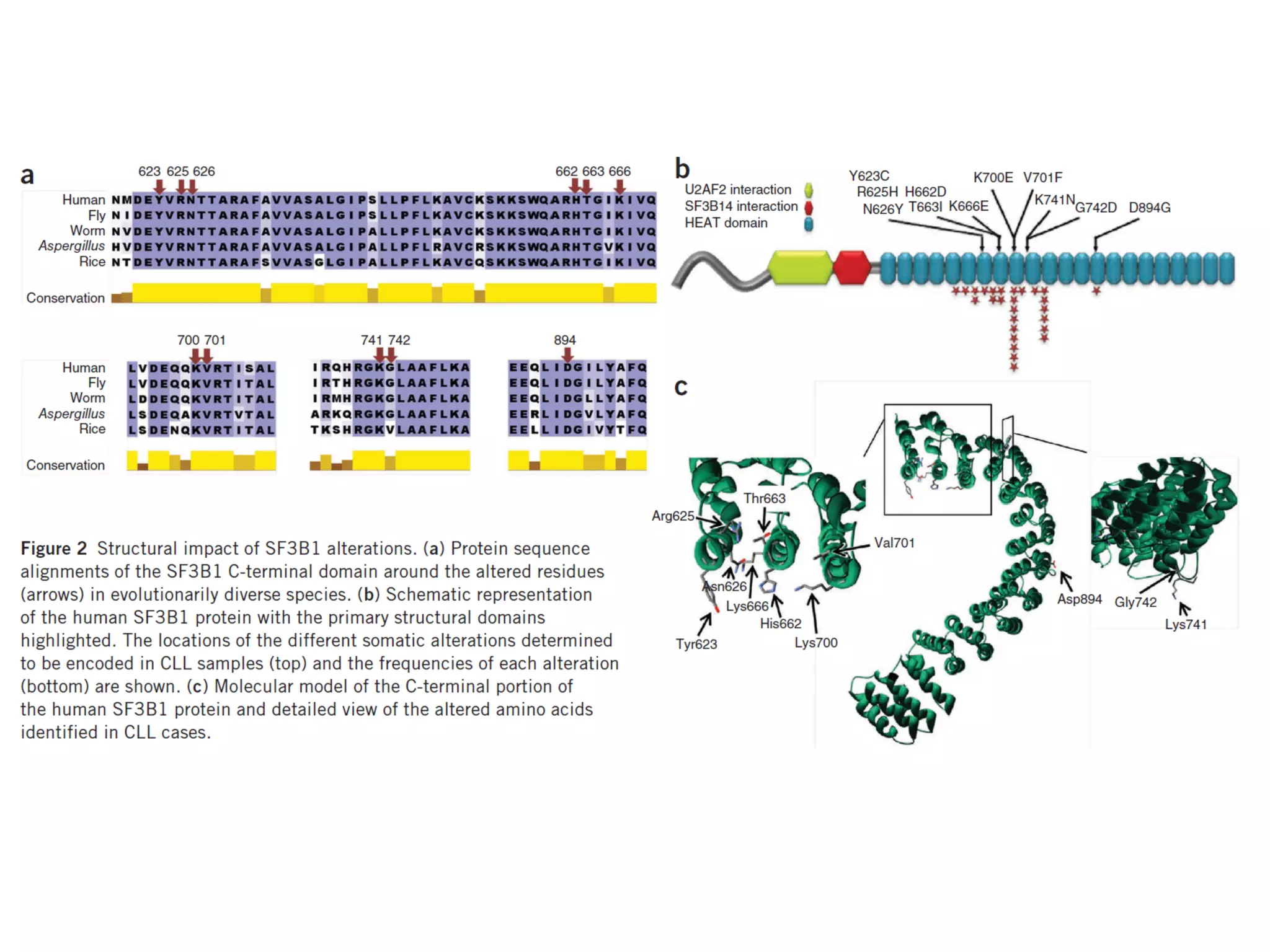
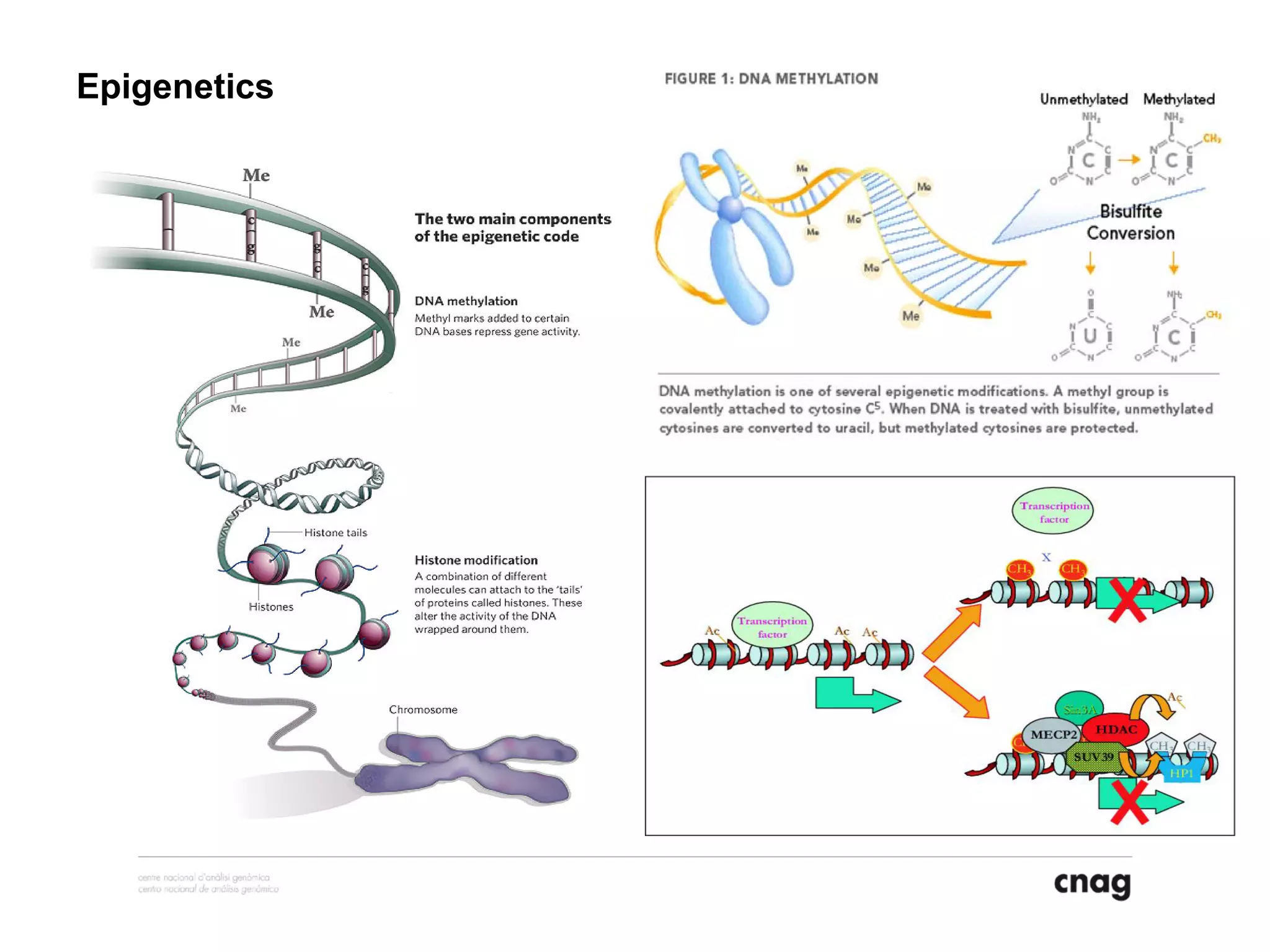
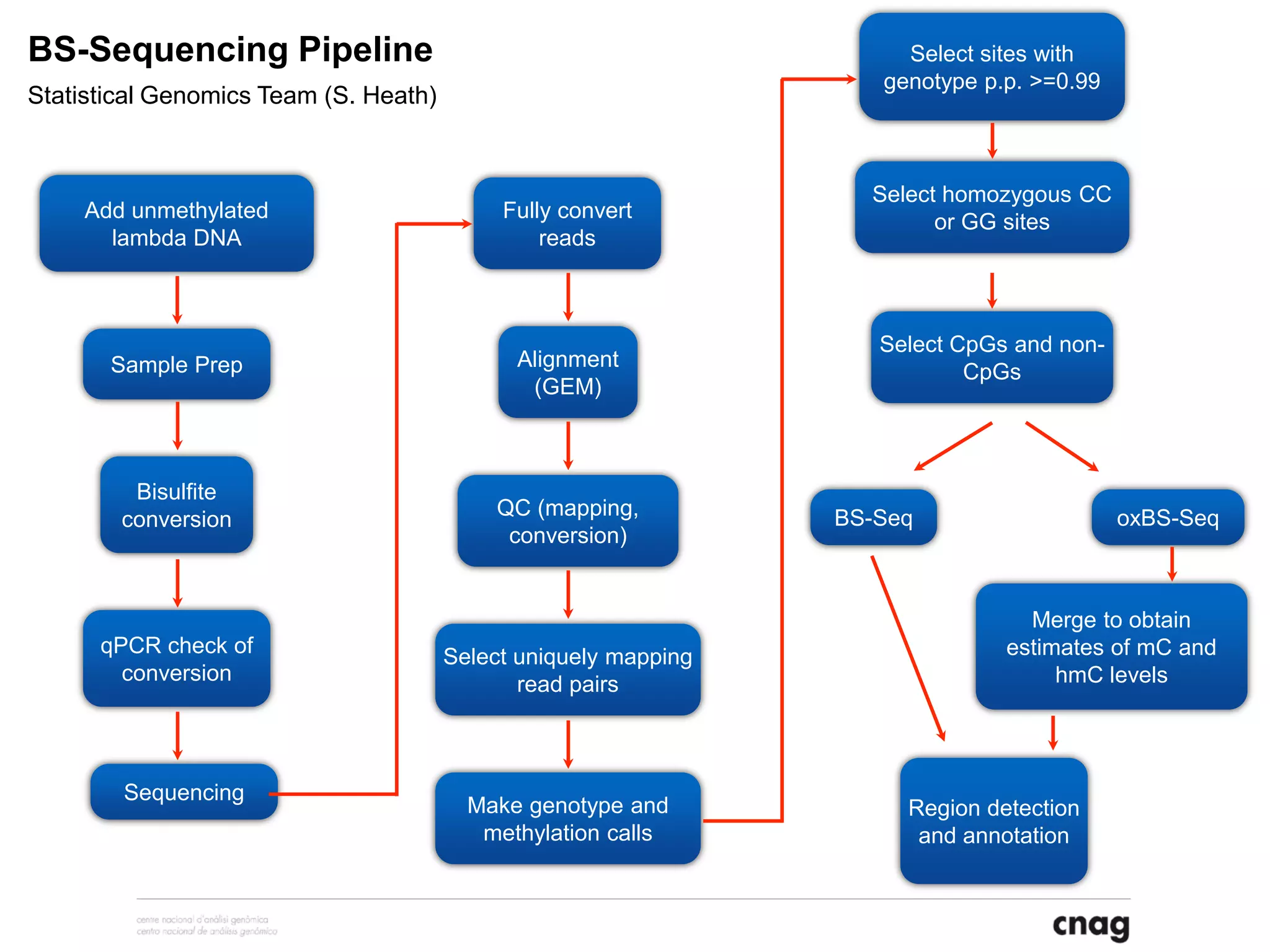
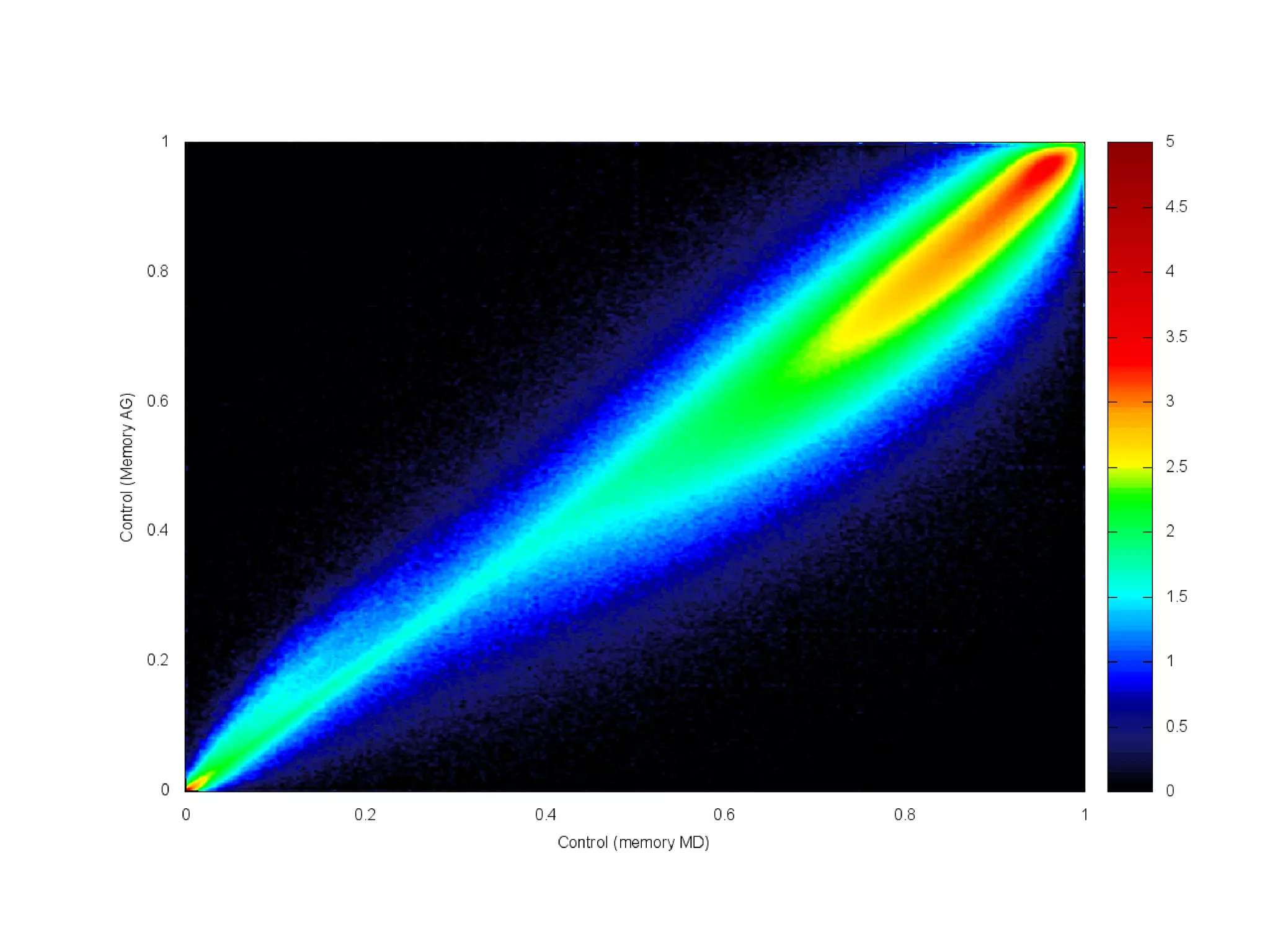
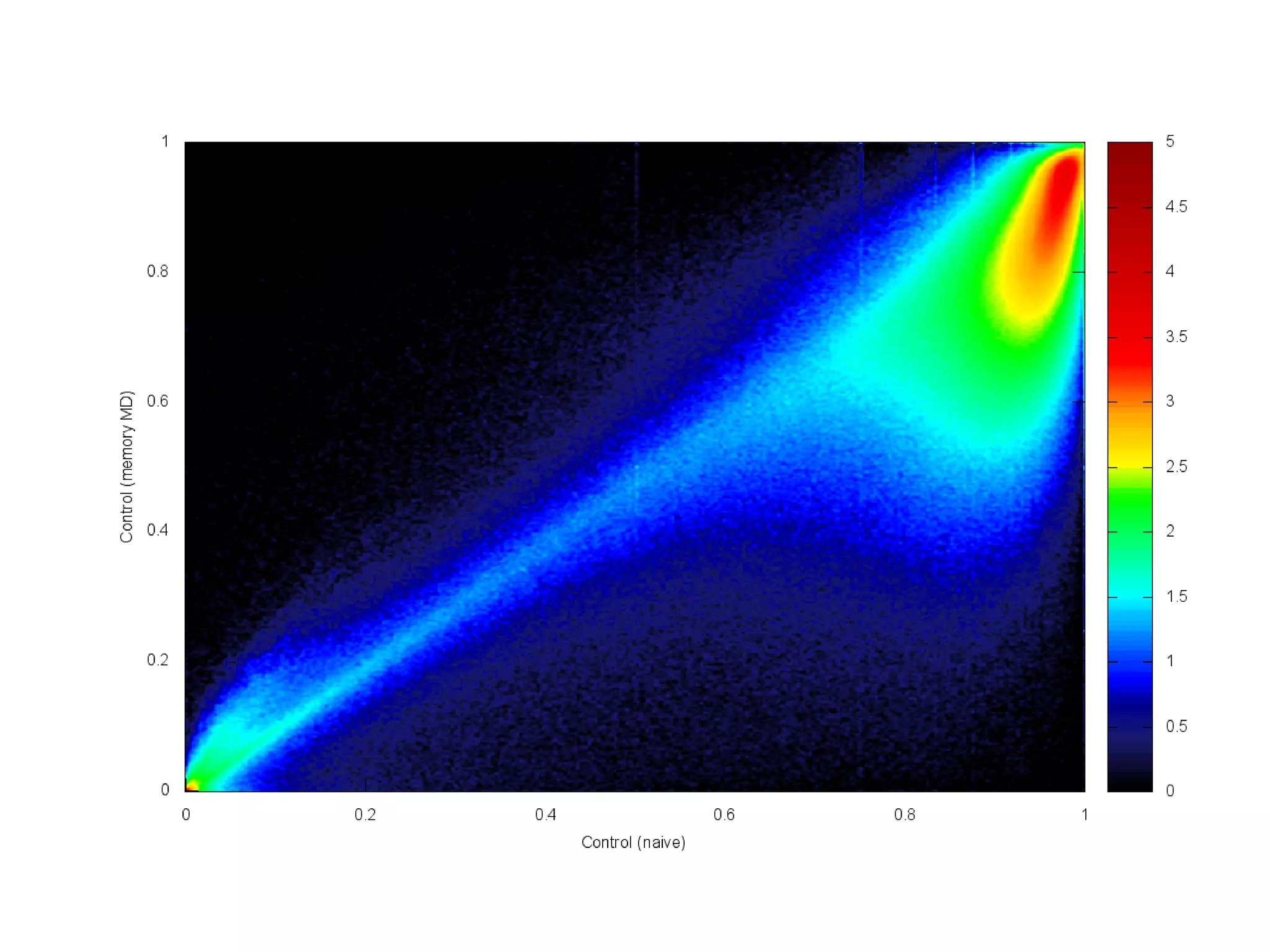
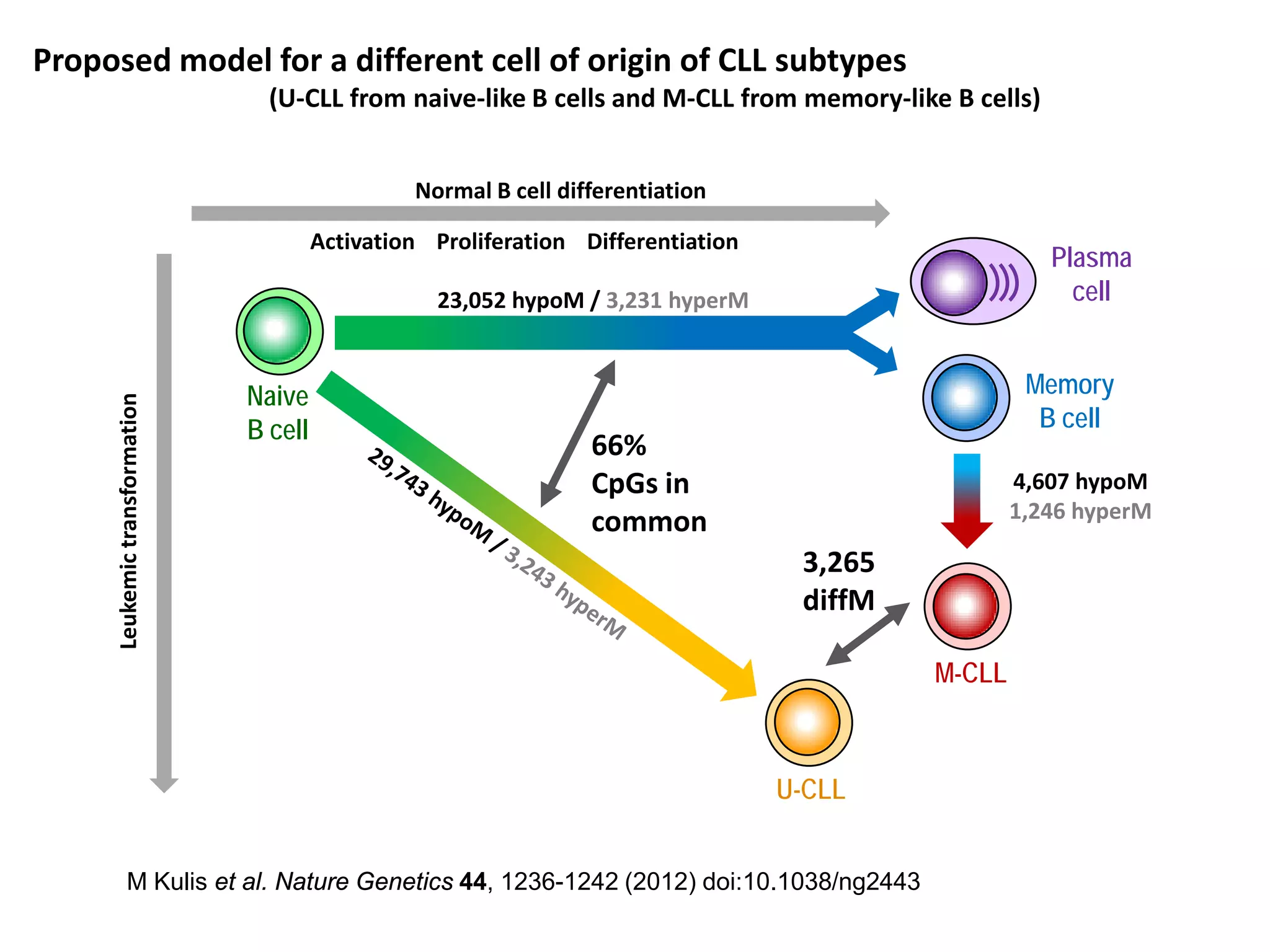
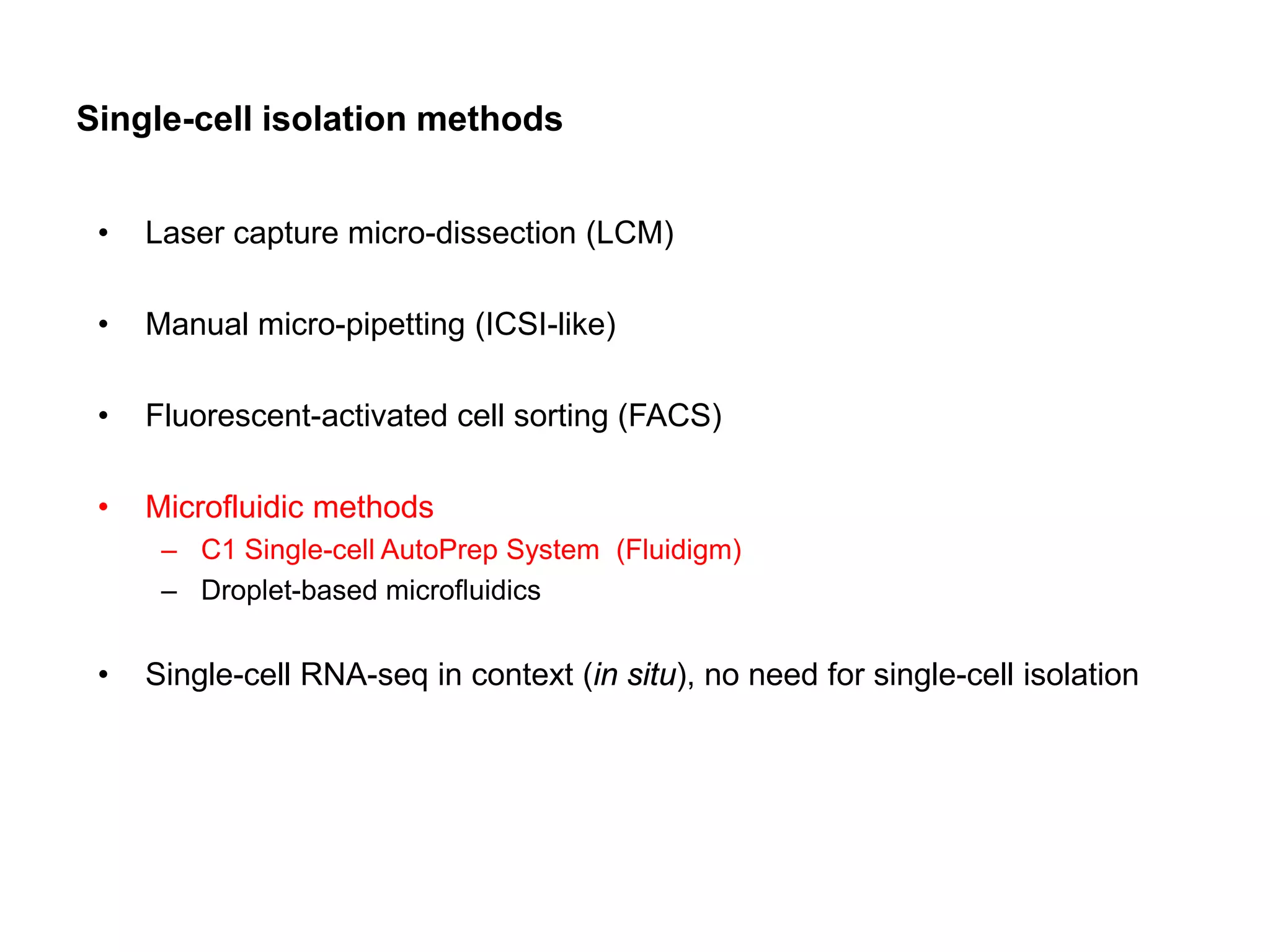
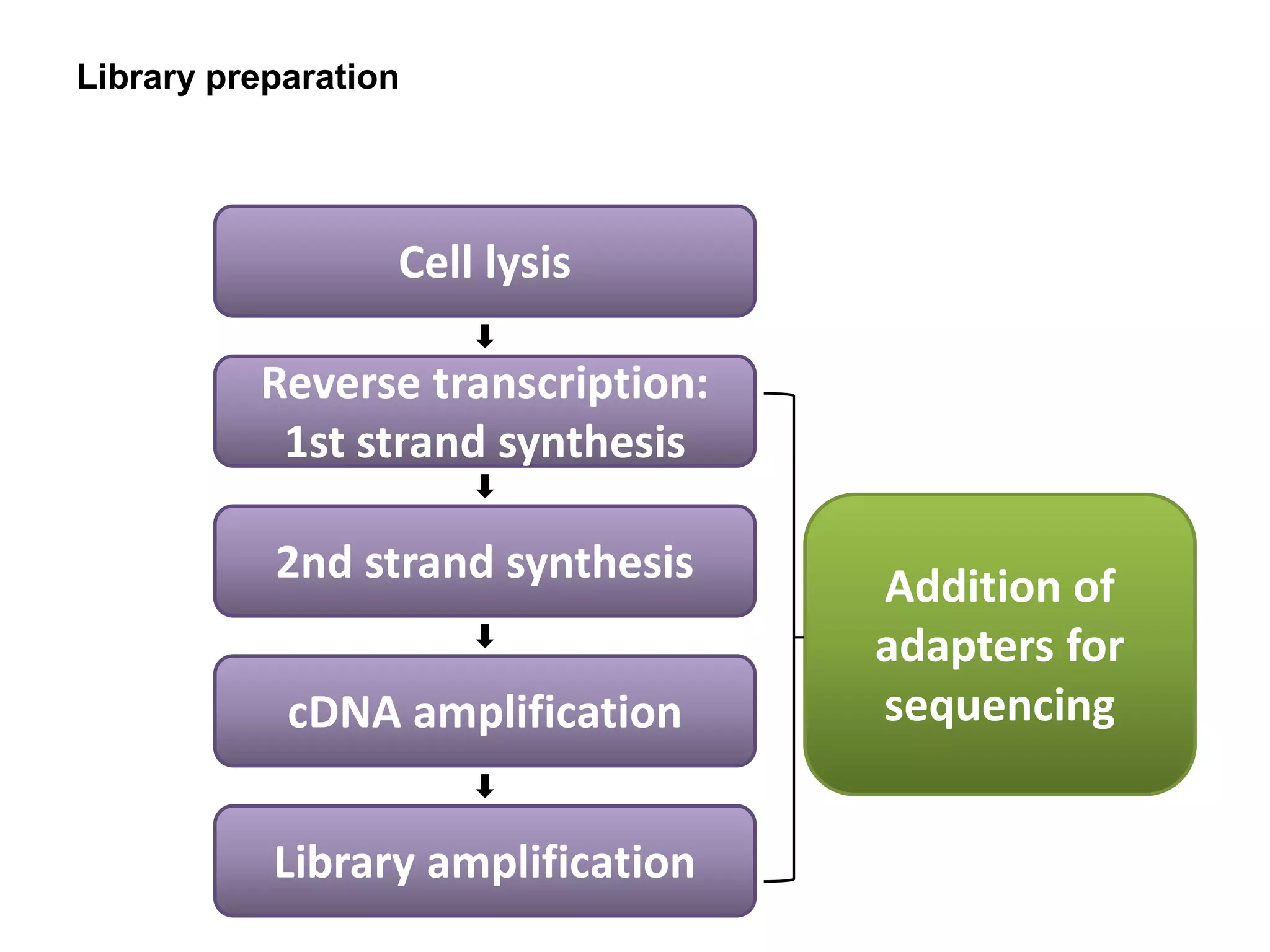
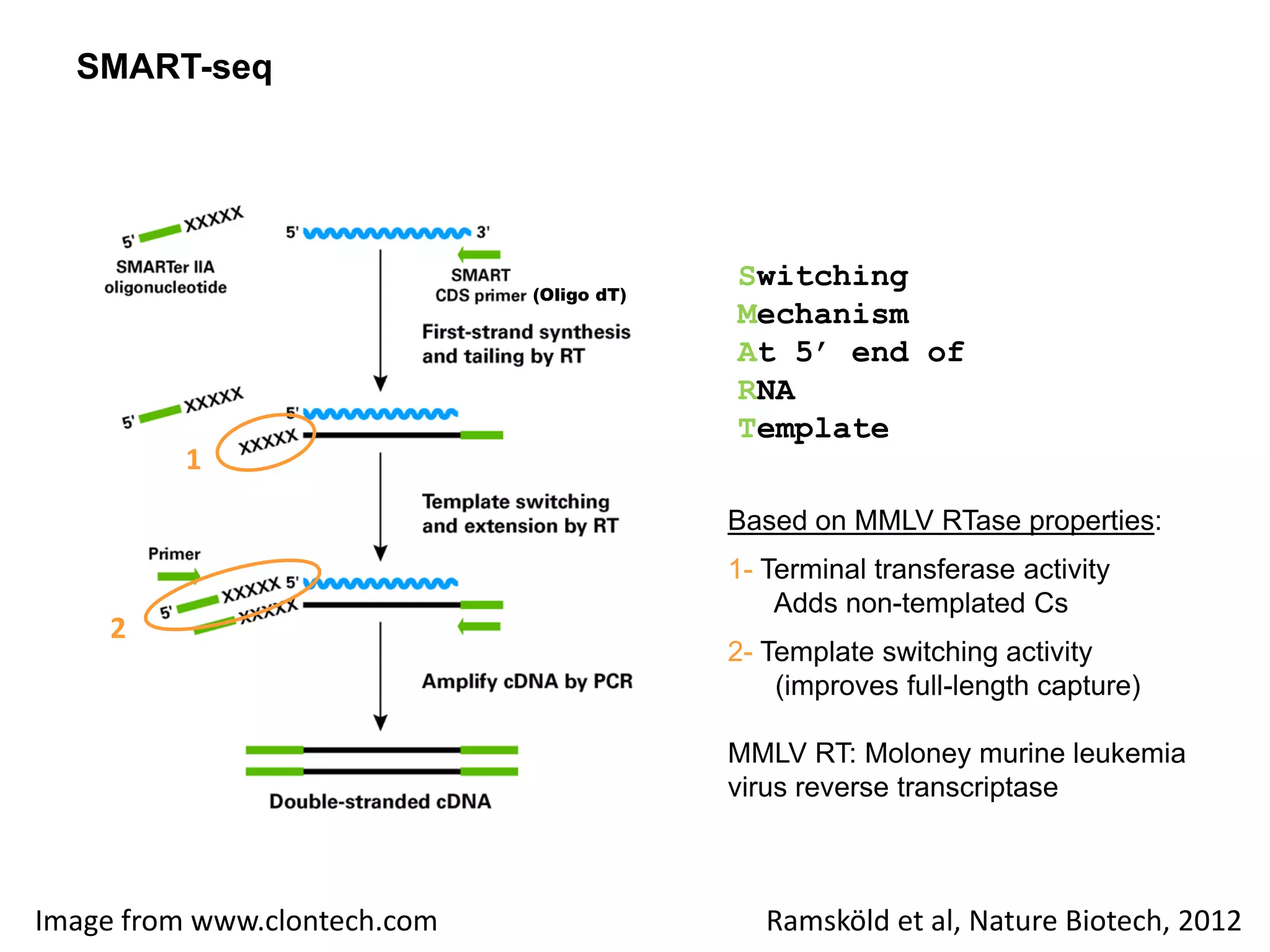
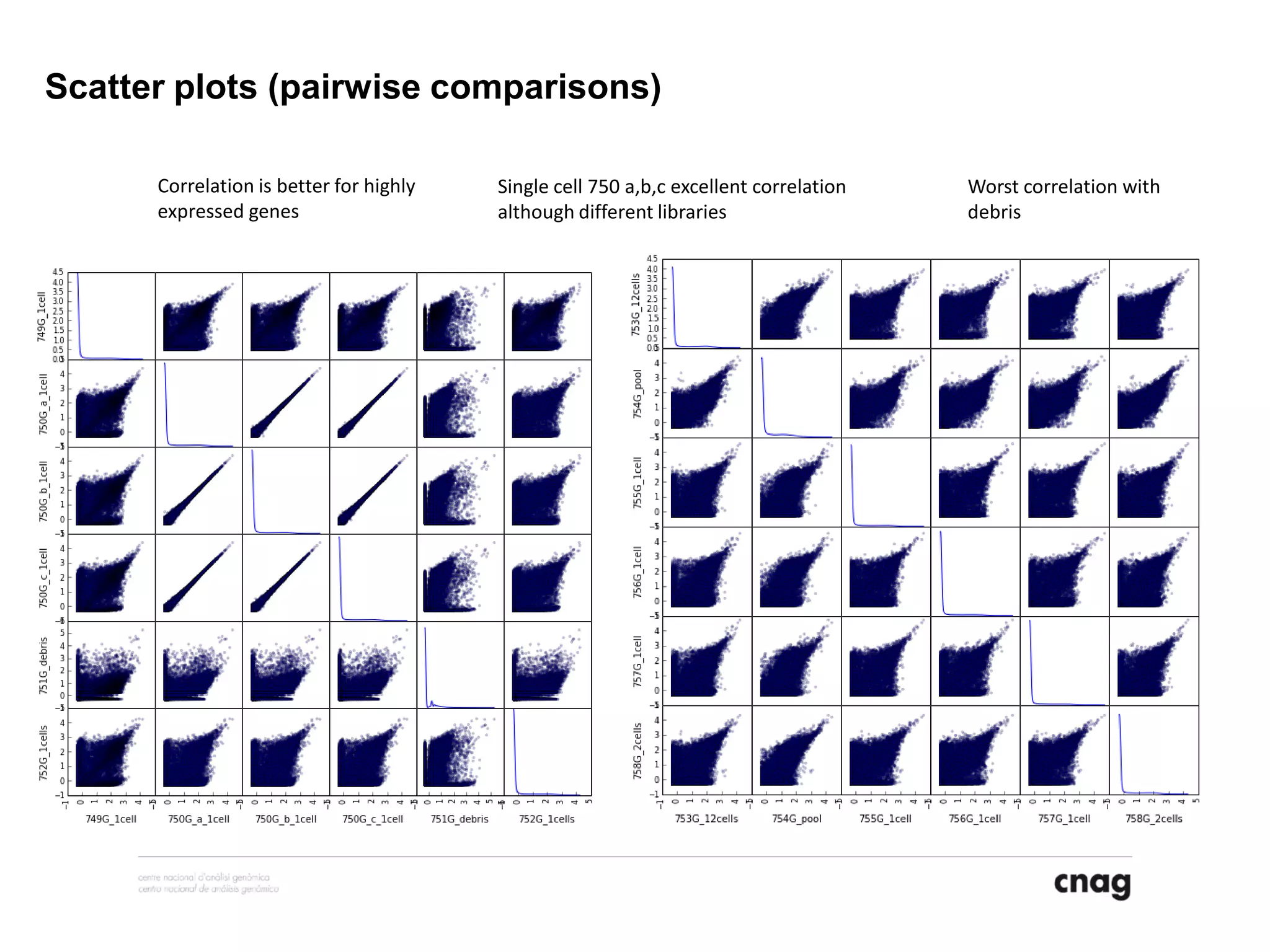
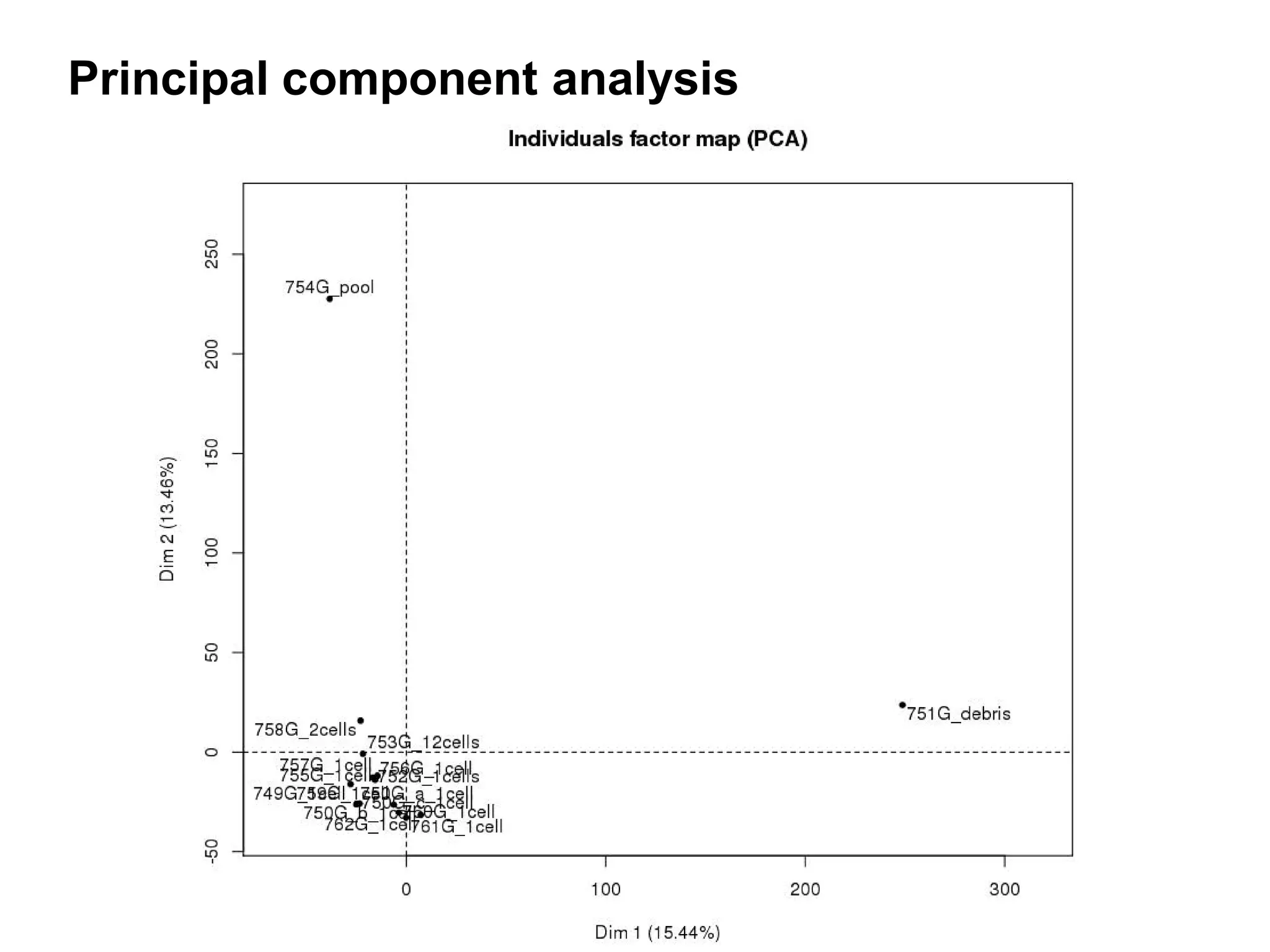
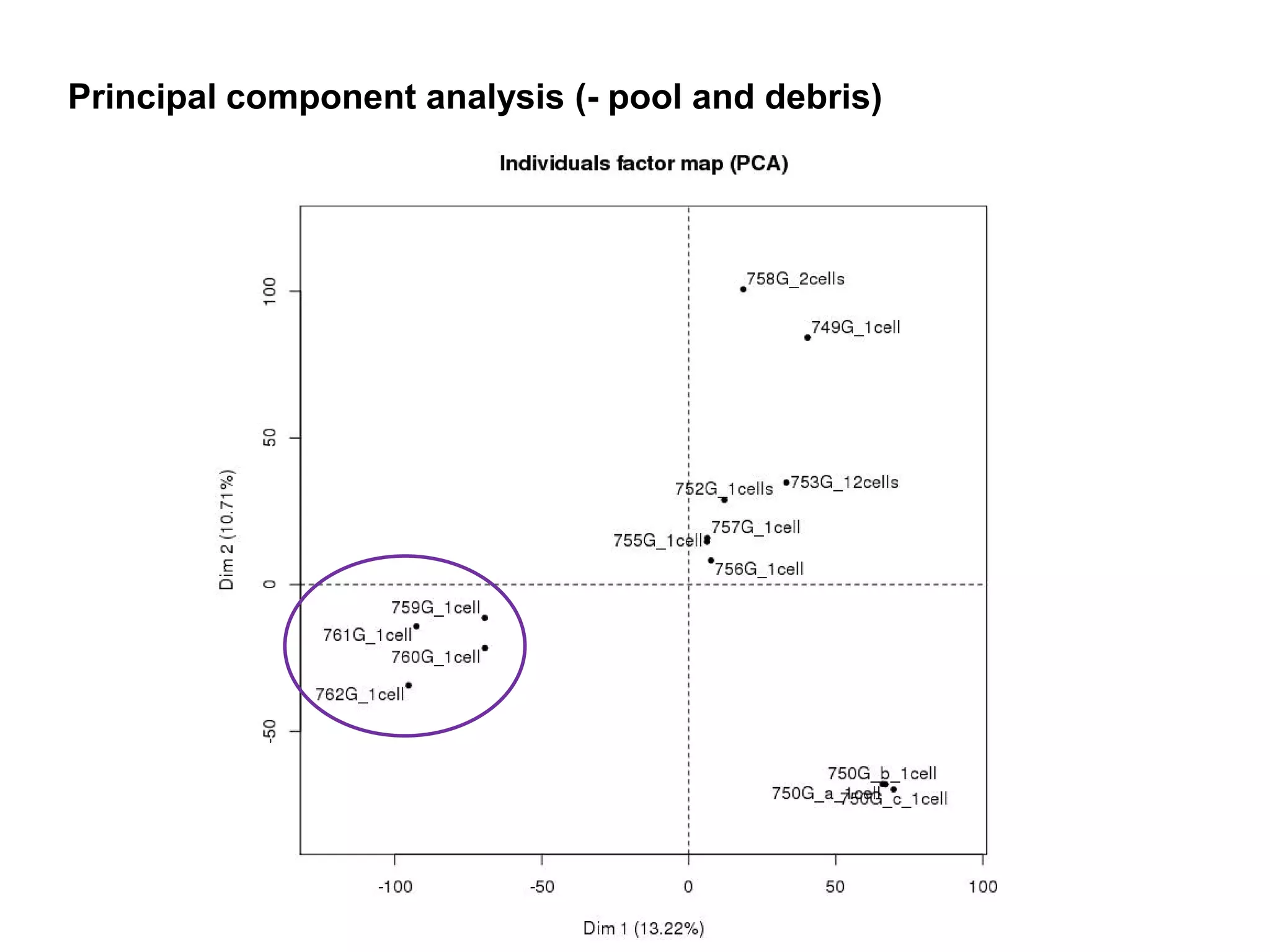
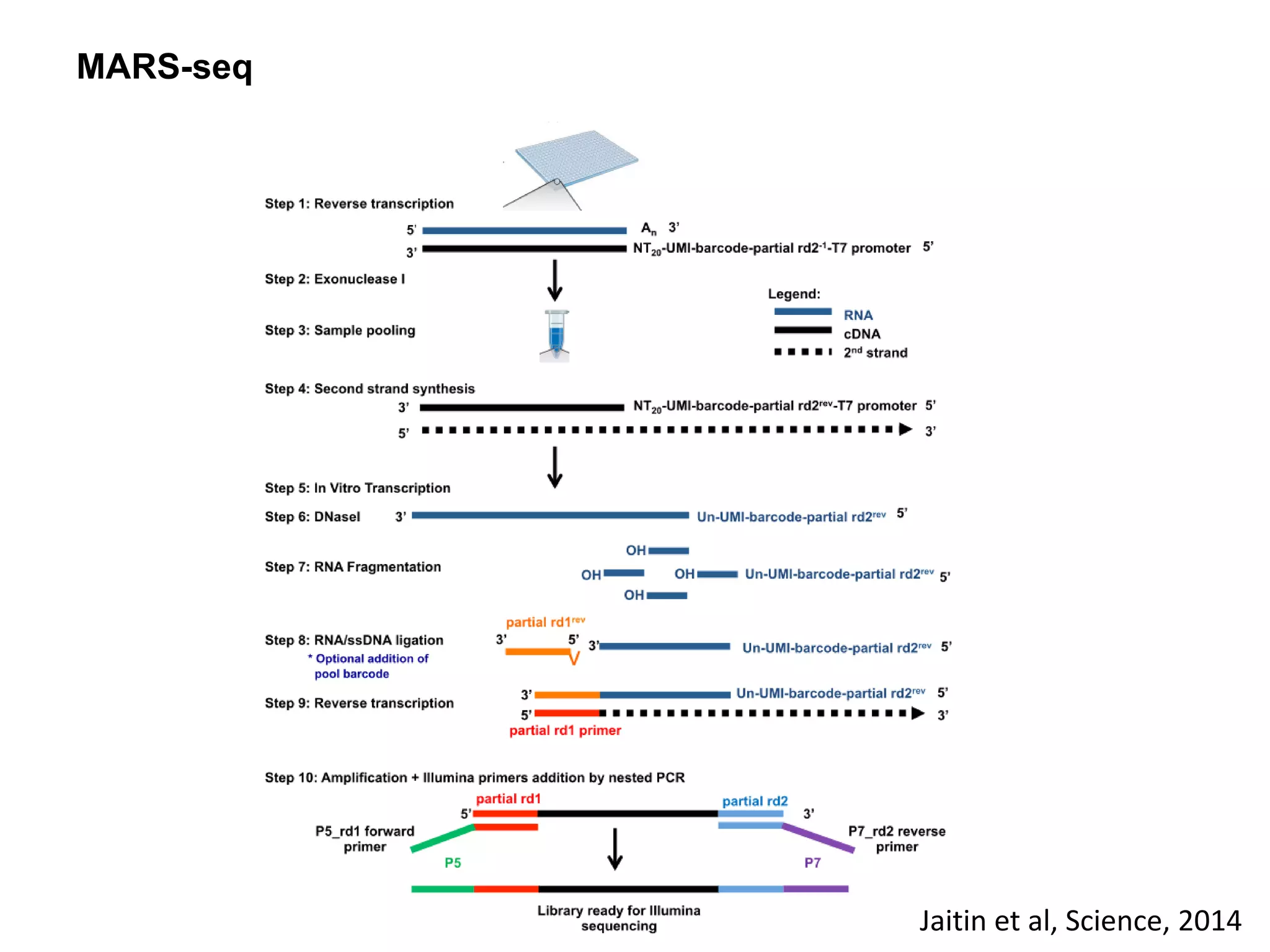
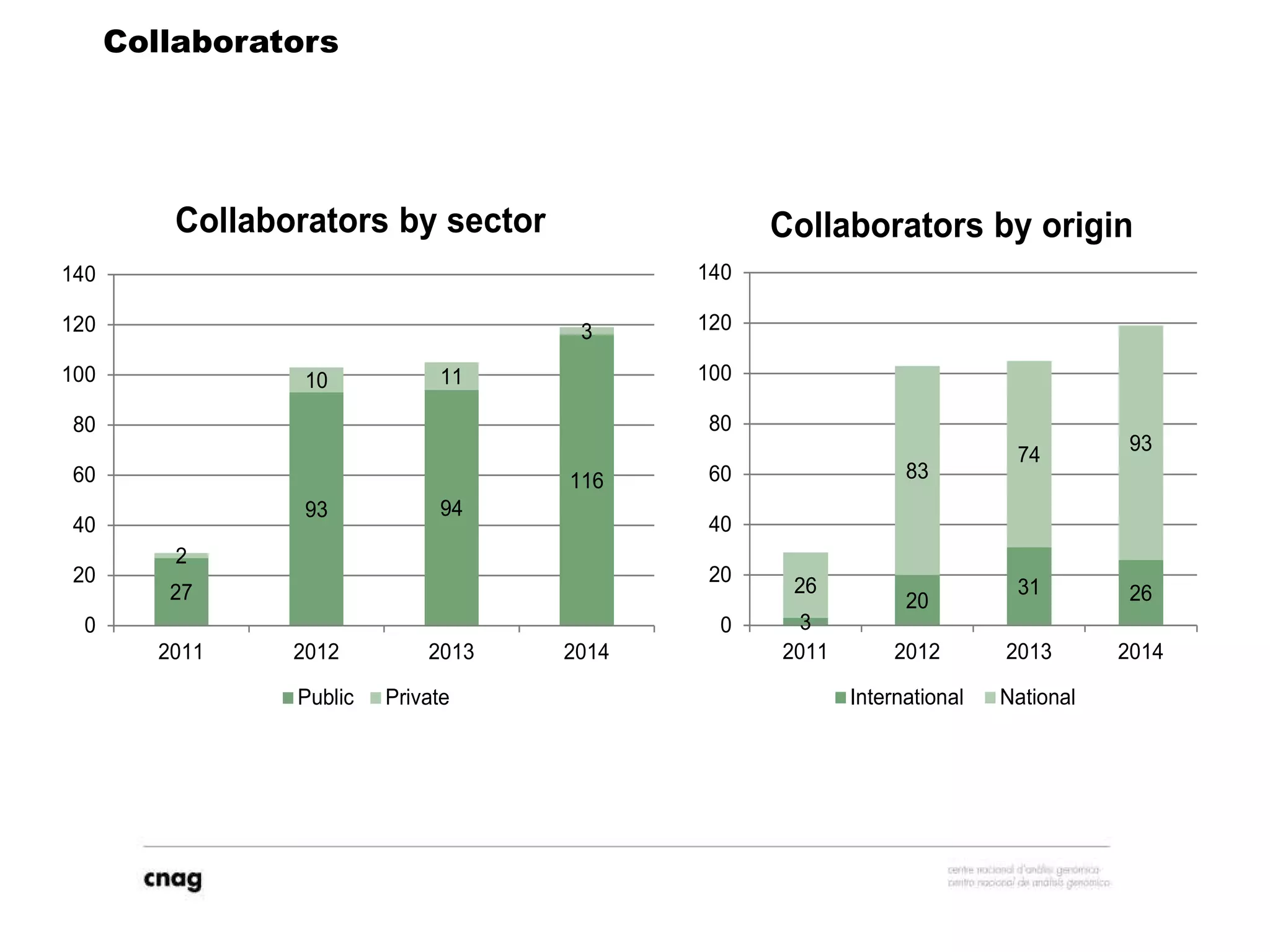
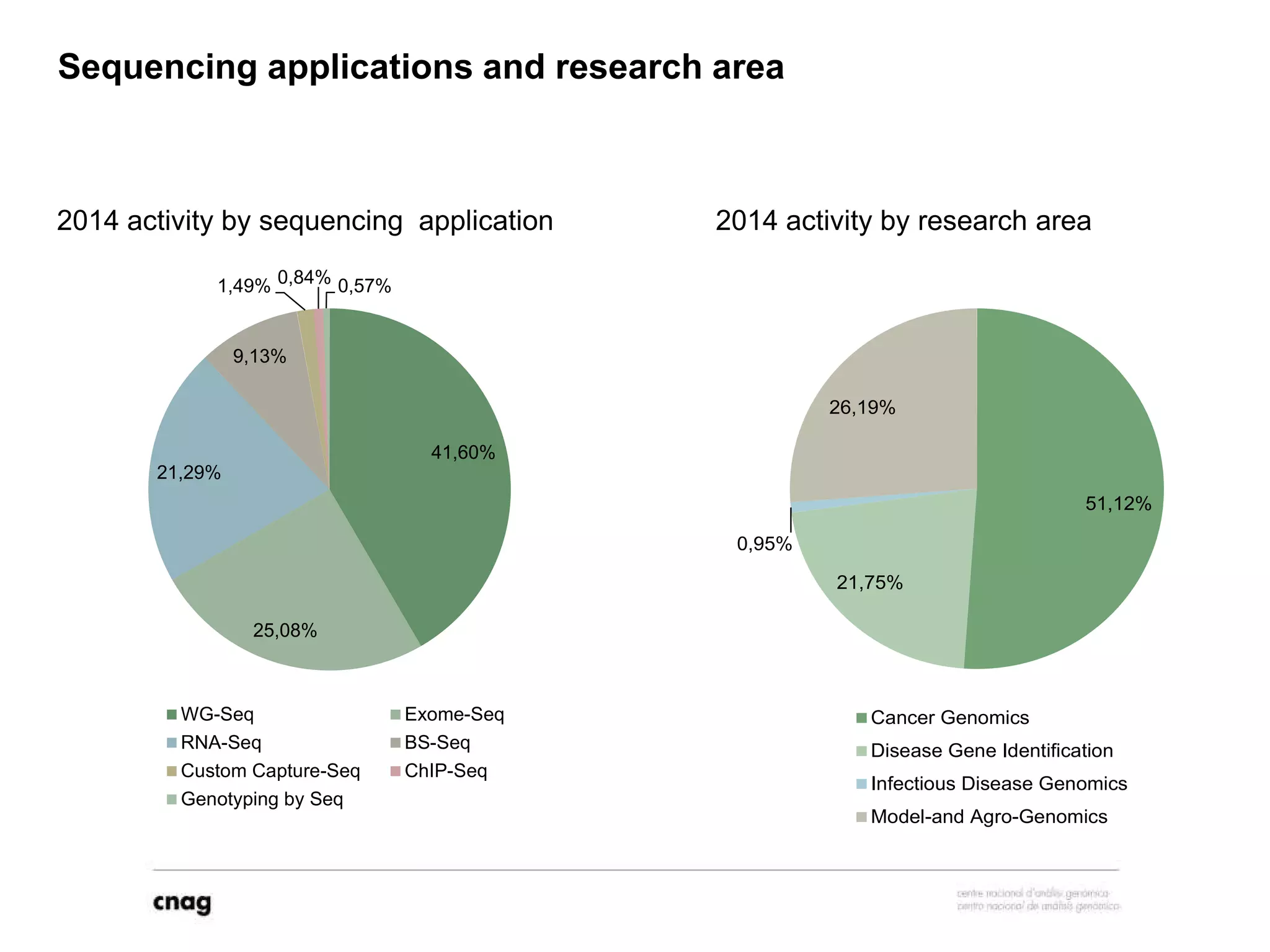
The document provides information about a short course on next generation sequencing and analysis of sequence variants. It includes an agenda with sessions on introduction to NGS applications in medical research, data analysis pipelines, interpretation of variants, and tools for predicting pathogenicity. It also provides background on the organizing institutions, the CNAG sequencing center and its projects, and an overview of bioinformatics analysis pipelines and resources.