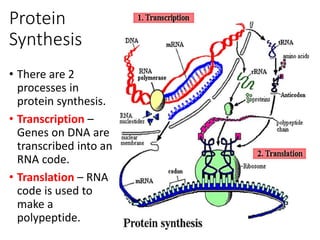
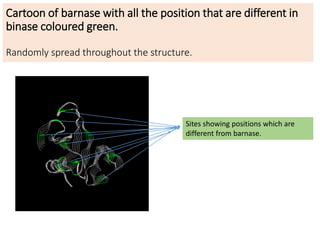
1) G.N. Ramachandran created the Ramachandran plot in 1963, which is an essential tool for understanding protein structure. The plot analyzes allowed regions of phi and psi dihedral angles in peptide units.
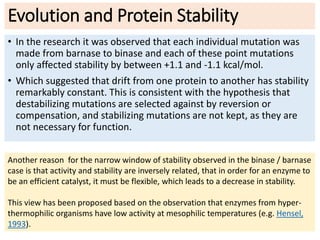
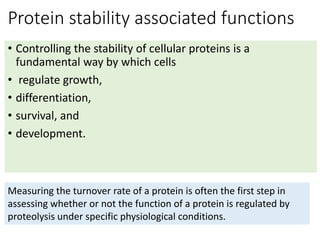
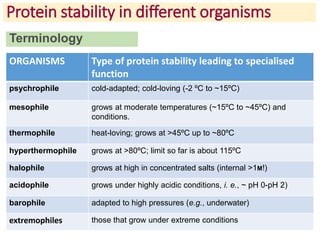
2) Protein stability refers to a protein maintaining its native folded conformation rather than becoming denatured. Stability depends on a balance of forces and is important for protein function.
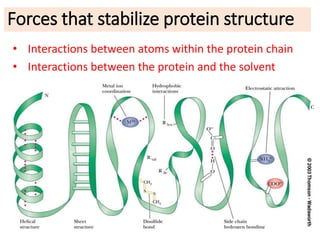
3) Factors that influence protein stability include pH, ligand binding, disulfide bonds, and interactions within the protein and between the protein and solvent. Chaperone proteins and proteases also help maintain stability in cells.