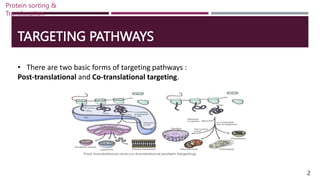
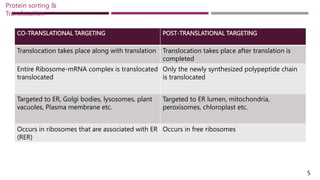
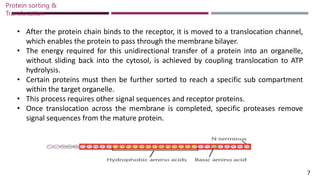
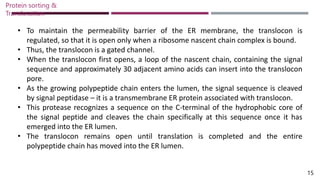
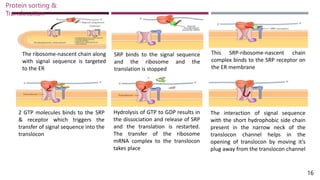
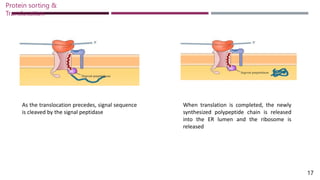
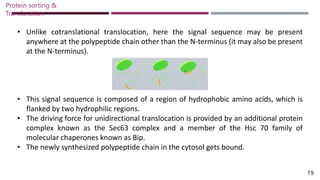
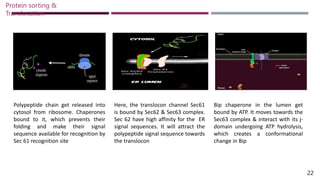
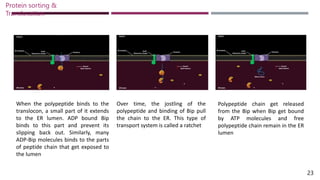
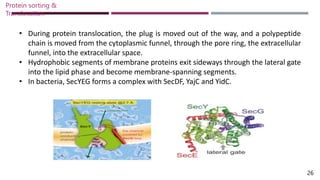
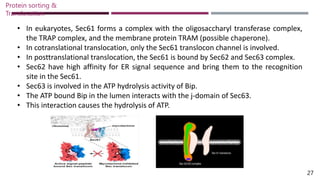
The document discusses protein sorting and translocation processes in mammalian cells, focusing on post-translational and co-translational targeting mechanisms. It highlights the roles of signal sequences and the signal recognition particle (SRP) in directing nascent proteins to their appropriate cellular locations, as well as the function of translocons in transporting these proteins across membranes. A detailed explanation of the energy requirements, protein interactions, and the processes involved in transporting proteins into the endoplasmic reticulum (ER) is provided.