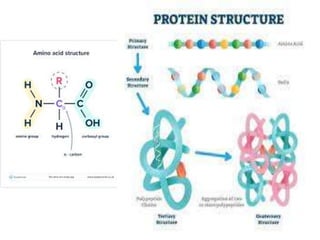
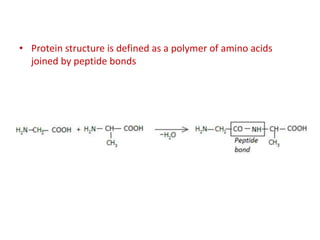
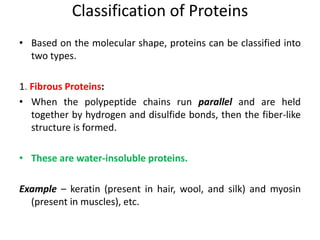
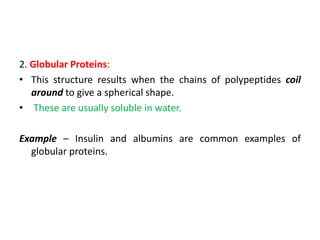
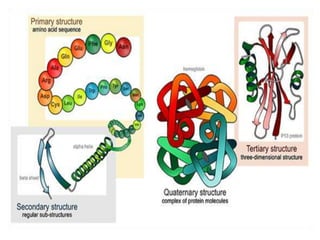
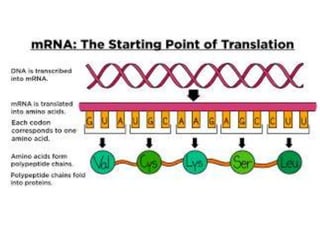
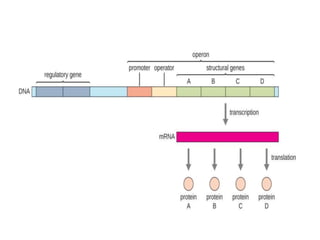
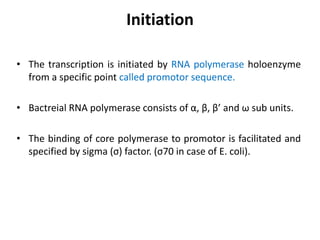
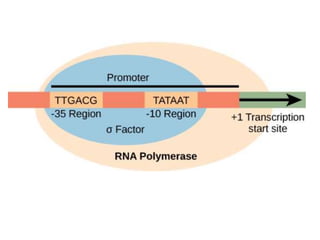
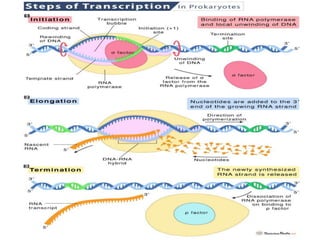
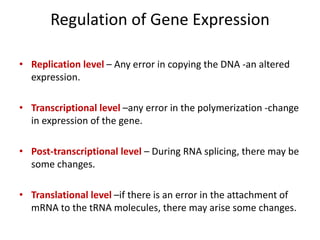
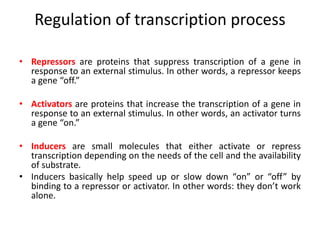
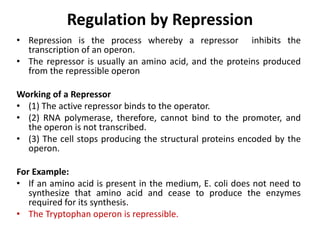
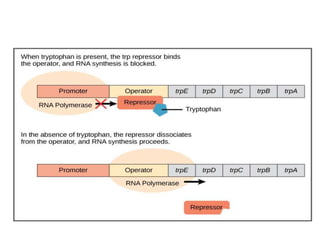
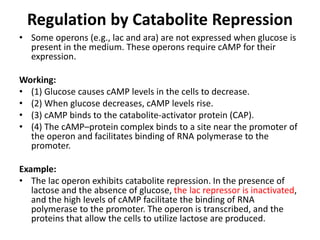
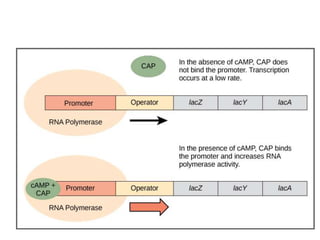
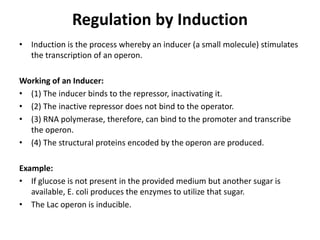
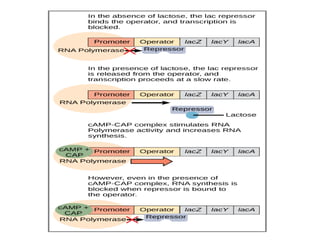
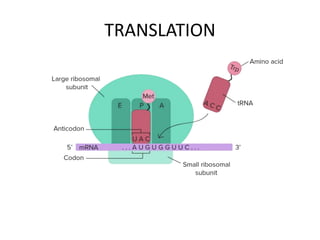
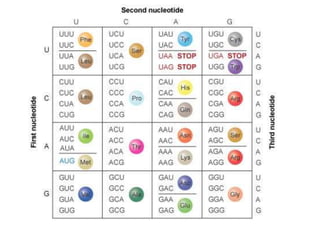
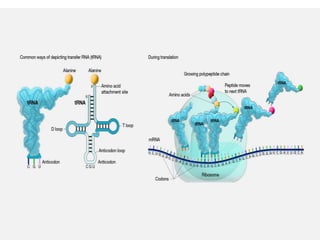
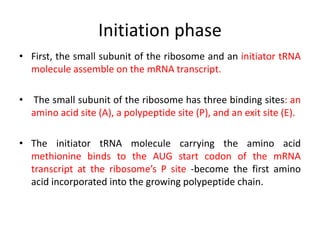
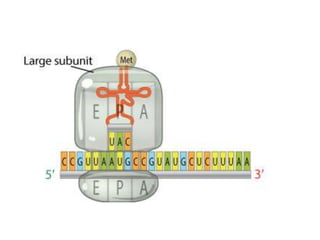
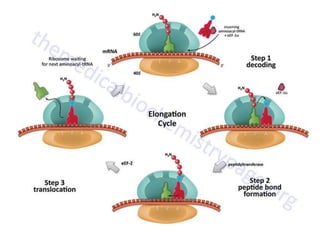
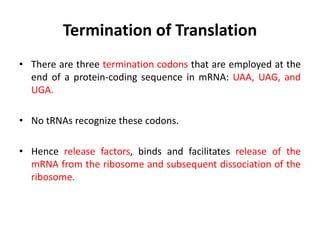
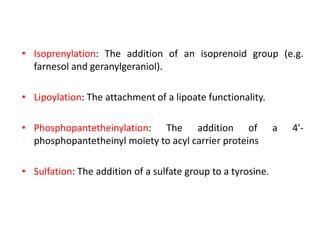
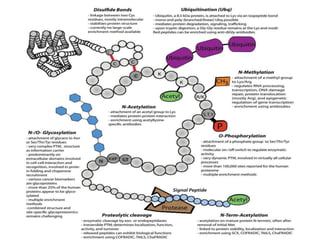
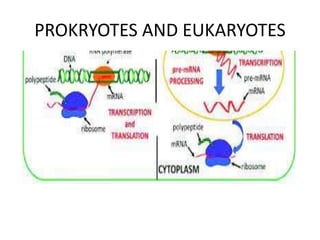
The document discusses protein synthesis in prokaryotes, detailing the structure and classification of proteins, and the processes of transcription and translation. It explains how genes are organized into operons and regulated by various mechanisms, including repressors, activators, and inducers. Additionally, it covers the steps of initiation, elongation, termination in transcription and translation, and highlights post-translational modifications that proteins undergo to become functional.