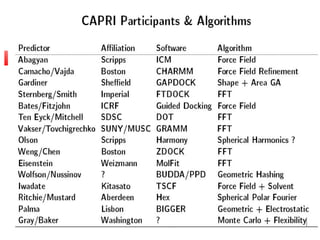
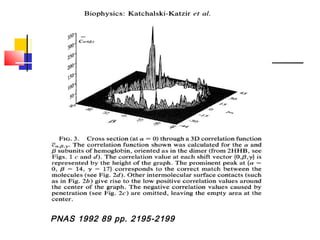
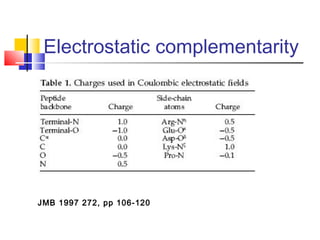
Protein docking is an important computational technique for understanding protein-protein interactions. It seeks to predict the binding orientation of two protein structures. Docking is a difficult problem due to the large search space of possible orientations and conformations. Successful docking requires scoring potential poses based on factors like free energy, buried surface area, hydrogen bonding, and complementarity of charges. Several search strategies are used like simulated annealing and genetic algorithms. International blind tests like CAPRI evaluate docking methods and provide lessons on better accounting for flexibility and active sites. Future challenges include improving scoring functions and models of flexibility.




![Force field
+−+−= ∑∑ 22
)()( eq
angle
eq
bonds
b krrkU θθθ
])[()]cos(1[
2 612
ij
ji
ij
ij
i ij ij
ij
dihedral n
n
r
qq
r
B
r
A
n
V
⋅
+−+−+ ∑∑∑ ∑ > ε
γφ
kb – bond parameter
kө --- angle parameter
Vn --- dihedral energy barrier
Van Der Waal radii
Partial charge set](https://image.slidesharecdn.com/mypresentation7-130430000428-phpapp01/85/Protein-docking-5-320.jpg)