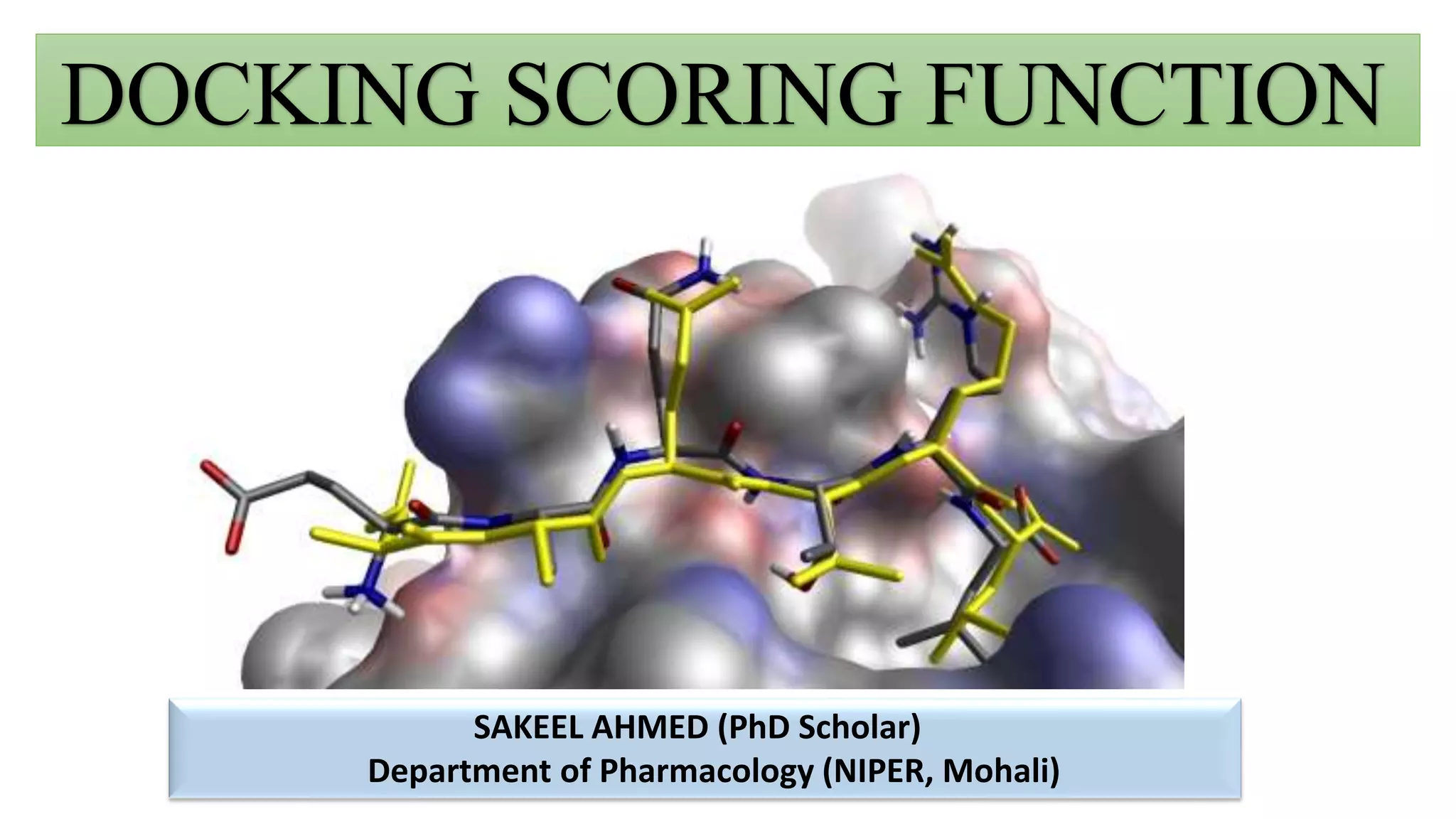
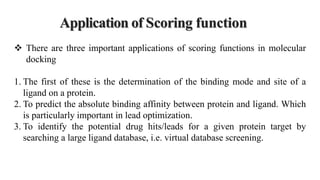
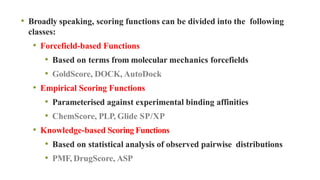
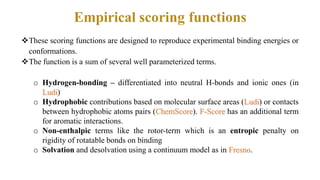
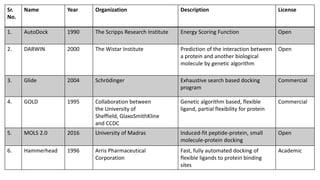
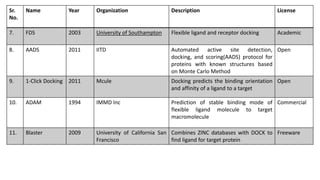
This document discusses docking scoring functions, which are mathematical functions used to predict the binding affinity between molecules after docking. There are three main applications of scoring functions: determining the binding mode of a ligand on a protein, predicting absolute binding affinity, and identifying potential drug hits through virtual screening. The document outlines different classes of scoring functions, including force field-based, empirical, knowledge-based, consensus, and shape/chemical complementary scores. It provides examples of popular docking programs that utilize different scoring function approaches.