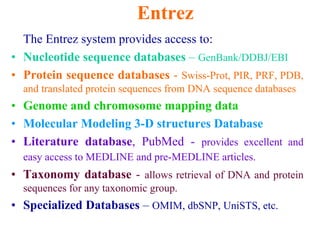
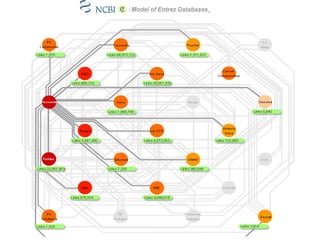
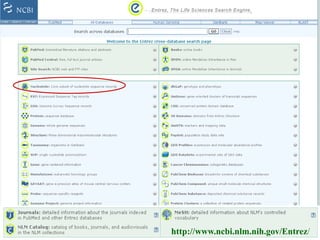
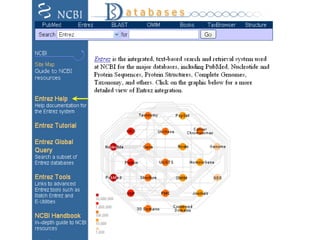
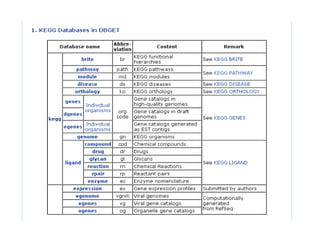
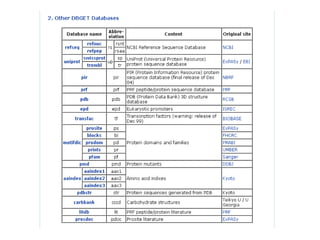
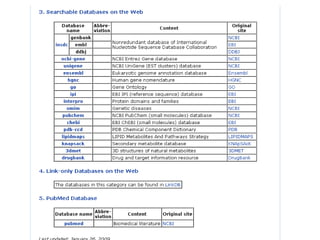
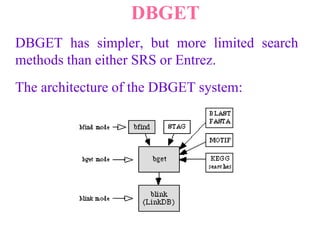
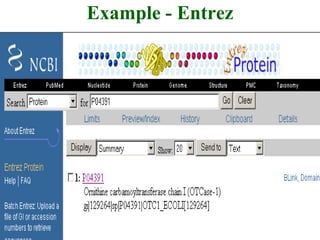
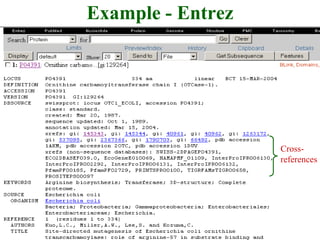
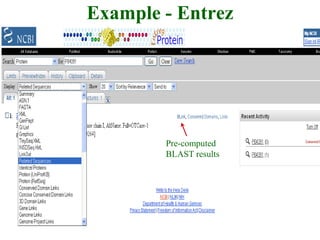
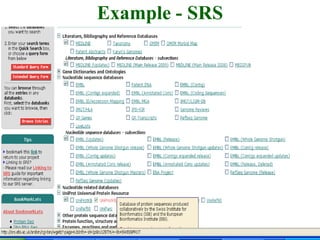
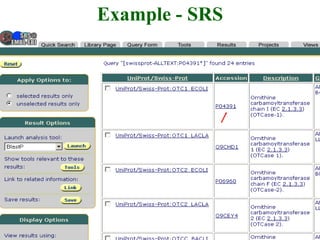
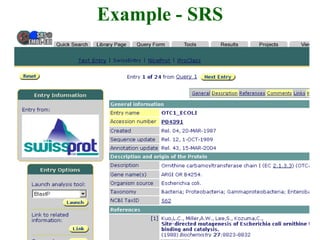
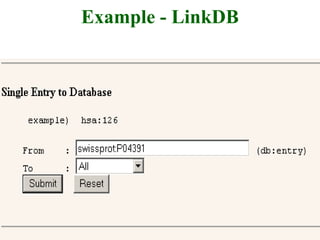
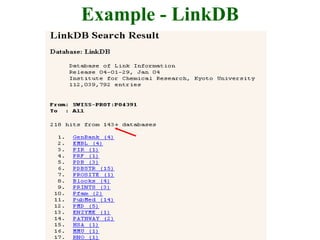
The document discusses different text-based database retrieval systems for accessing biological data, including Entrez, SRS, and DBGET/LinkDB. It describes their key features and how each system allows users to search text databases using queries, with Entrez providing linked related data across multiple databases. An example shows how each system can be used to retrieve and view related information for a SwissProt protein entry.