Embed presentation
Downloaded 15 times










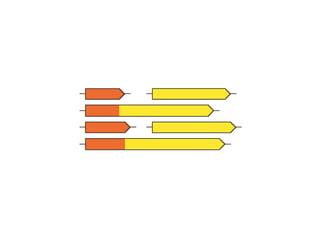

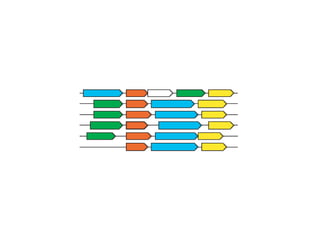

































![Gene and protein names Cue words for entity recognition Verbs for relation extraction [ nxgene The GAL4 gene ] [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ]](https://image.slidesharecdn.com/jen06talk9-1217971178211118-9/85/The-STRING-database-57-320.jpg)
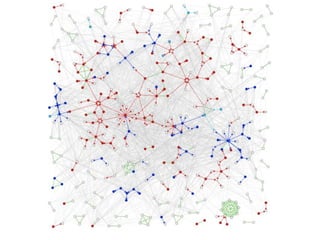



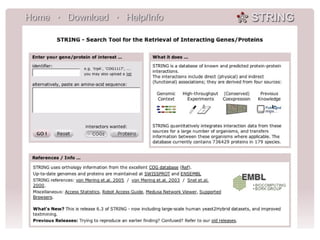

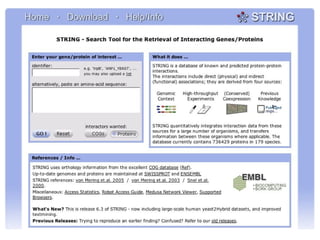

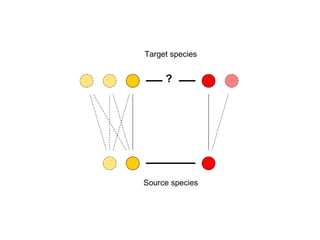

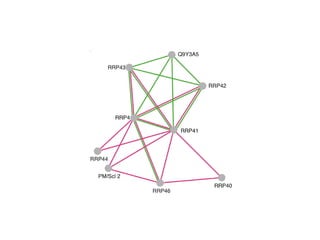


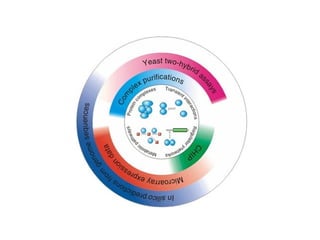
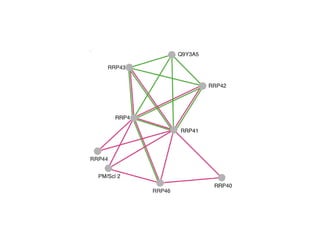
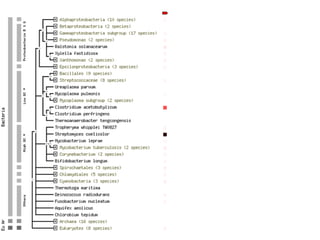
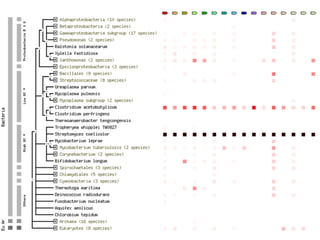
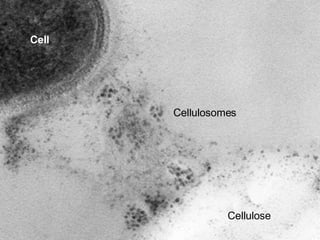
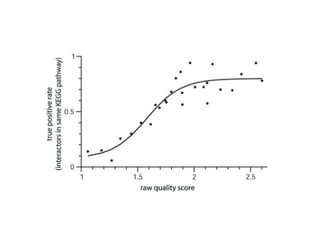
The document discusses the STRING database, which integrates various biological data sources to analyze functional interactions across 179 proteomes. It highlights methods for gene and protein interactions, including benchmarking against existing data and literature mining techniques. Acknowledgments are given to the STRING team and contributors for their work on the database and methodologies used.























































![Gene and protein names Cue words for entity recognition Verbs for relation extraction [ nxgene The GAL4 gene ] [ nxexpr T he expression of [ nxgene the cytochrome genes [ nxpg CYC1 and CYC7 ]]] is controlled by [ nxpg HAP1 ]](https://image.slidesharecdn.com/jen06talk9-1217971178211118-9/85/The-STRING-database-57-320.jpg)











