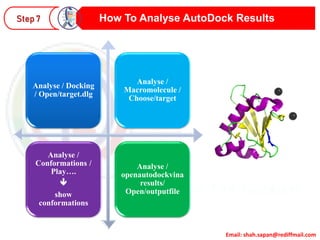
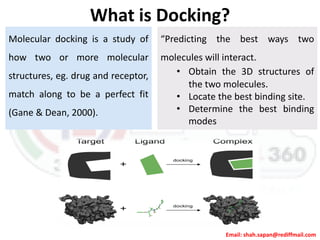
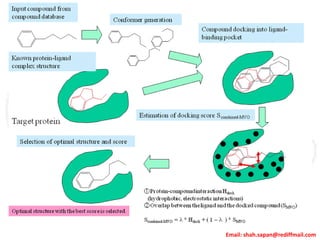
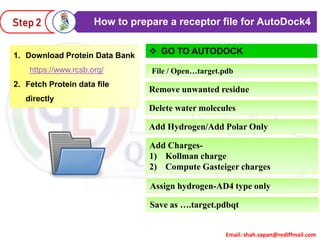
This document provides instructions for performing molecular docking simulations using Autodock 4. It discusses downloading and installing Autodock, preparing receptor and ligand files in pdbqt format, setting up a grid parameter file to define the docking grid, preparing an Autodock parameter file to define the docking parameters, running Autodock to perform the docking simulation, and analyzing the results to view and evaluate the predicted binding modes.







![Email: shah.sapan@rediffmail.com
How To Run AutoDock?Step 6
Run Autogrid
•Select tab RUN>>RUN AUTOGRID>>Click BROWSE>>Select AUTOGRID APPLICATION FILE in PROGRAM PATH
NAME>>Click BROWSE>>Select TARGET.GPF in PARAMET FILE NAME>>Click LAUNCH
Run Autodock
•Select tab RUN>>RUN AUTODOCK>>Click BROWSE>>Select AUTODOCK APPLICATION FILE in PROGRAM PATH
NAME>>Click BROWSE>>Select TARGET.DPF in PARAMET FILE NAME>>Click LAUNCH
Run Autodock Vina
• OPEN COMMAND PROMPT
• Type “C: foldername“ for selecting path
• Type "C:Program Files (x86)The Scripps Research InstituteVinaVina.exe" --
receptor target.pdbqt --ligand ligand.pdbqt --config conf.txt --log log.txt --out
output.pdbqtxt]](https://image.slidesharecdn.com/autodock-200427142126/85/Molecular-Docking-Using-Autodock-11-320.jpg)