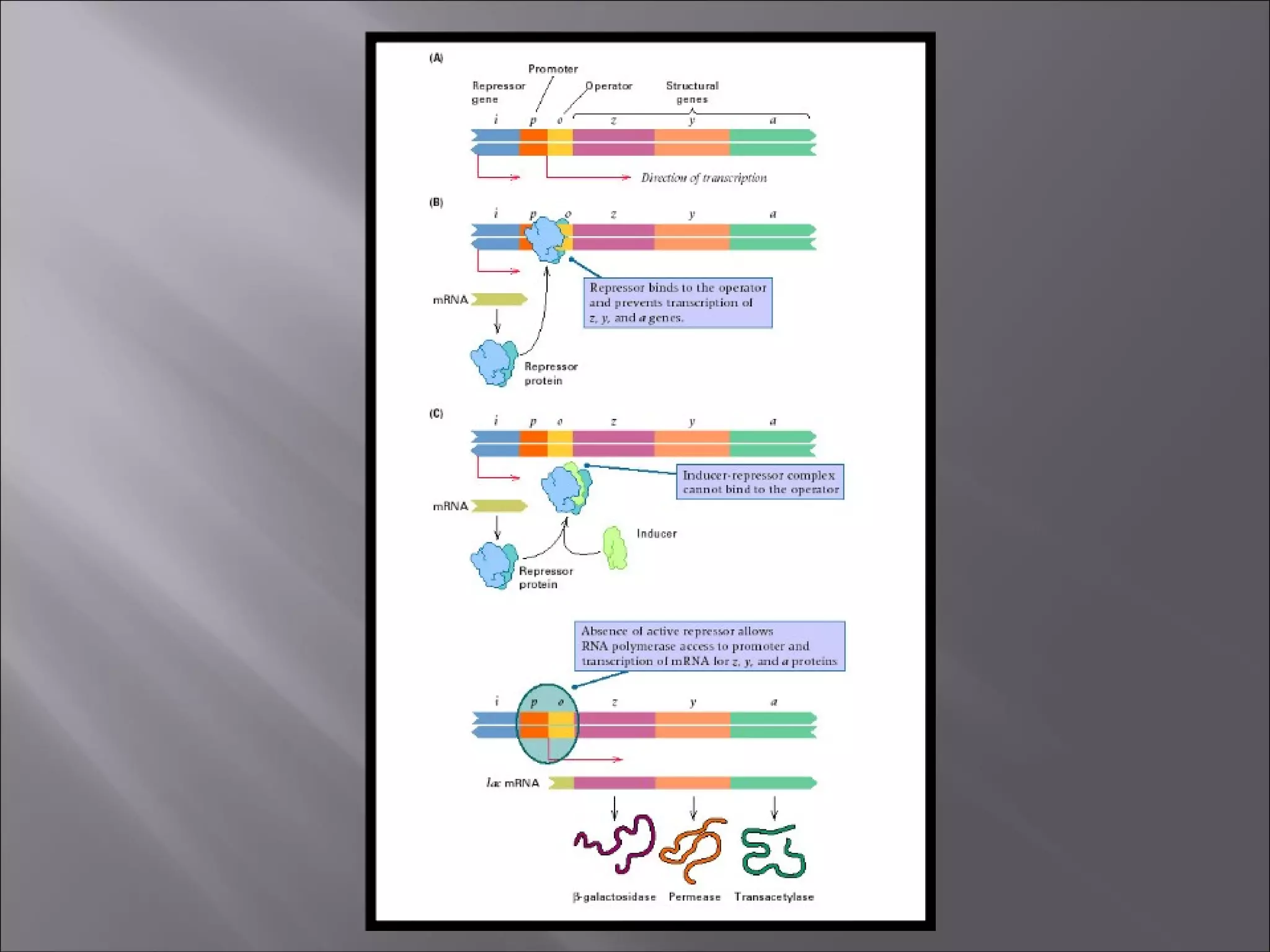
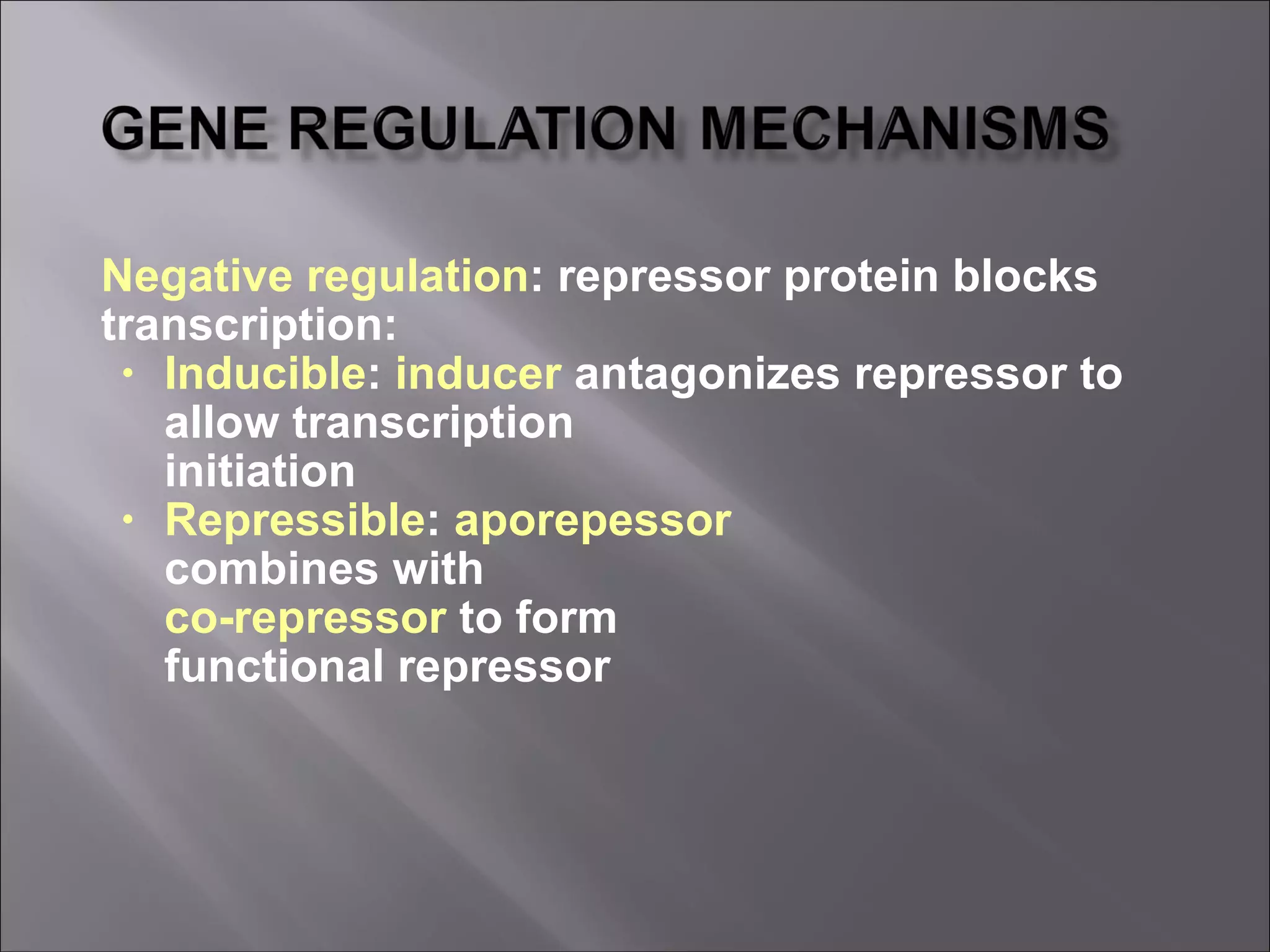
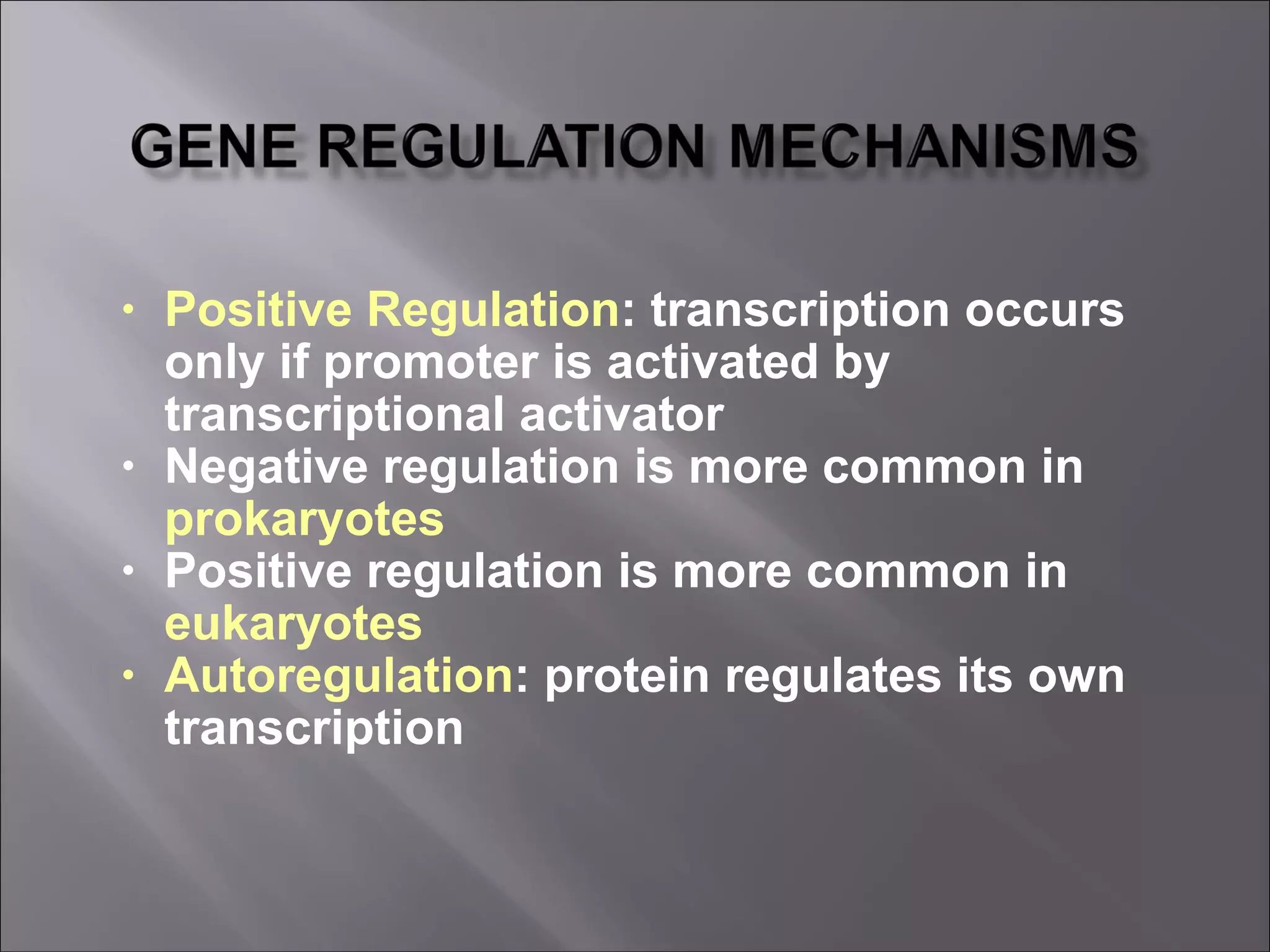
There are two main mechanisms of transcriptional control of gene expression in prokaryotes: 1) Operons, where genes involved in a metabolic pathway are clustered under common regulatory sequences and proteins, allowing synchronized expression. 2) Cascades of gene expression, where expression of one gene set can activate expression of another gene set. The lactose and tryptophan operons are examples that demonstrate negative and positive regulation of gene expression through repressors, inducers, and feedback loops in response to environmental conditions.