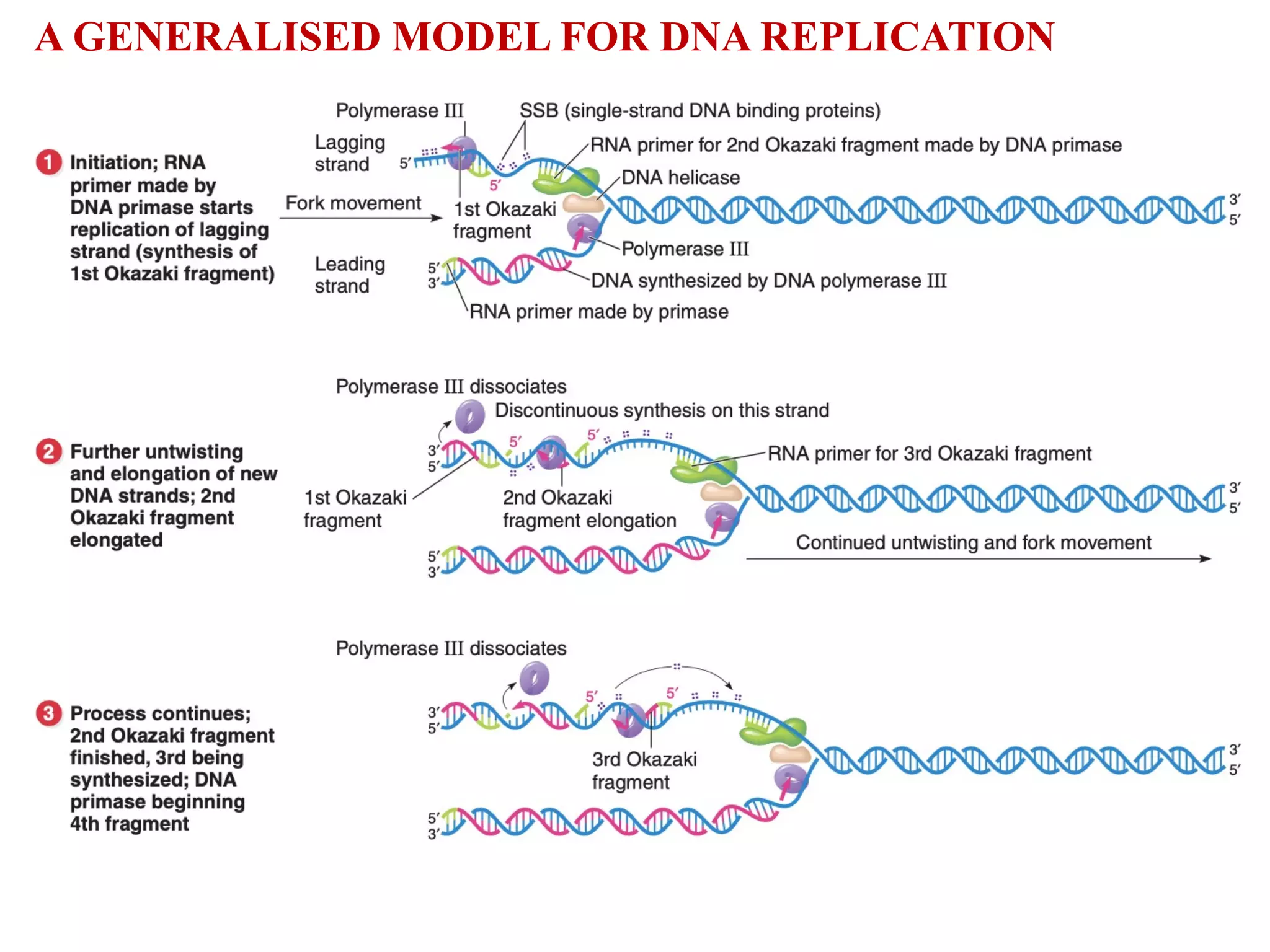
The document discusses DNA replication, defining it as the process through which a DNA molecule makes identical copies, with the semiconservative model being the widely accepted theory. It details the evidence from Meselson and Stahl's experiment using isotopes to demonstrate this model, highlighting the formation of distinct DNA bands after cell generations. Additionally, the document outlines the roles of various enzymes involved in DNA replication and the concepts of replicons, origin, and terminus in both prokaryotic and eukaryotic cells.