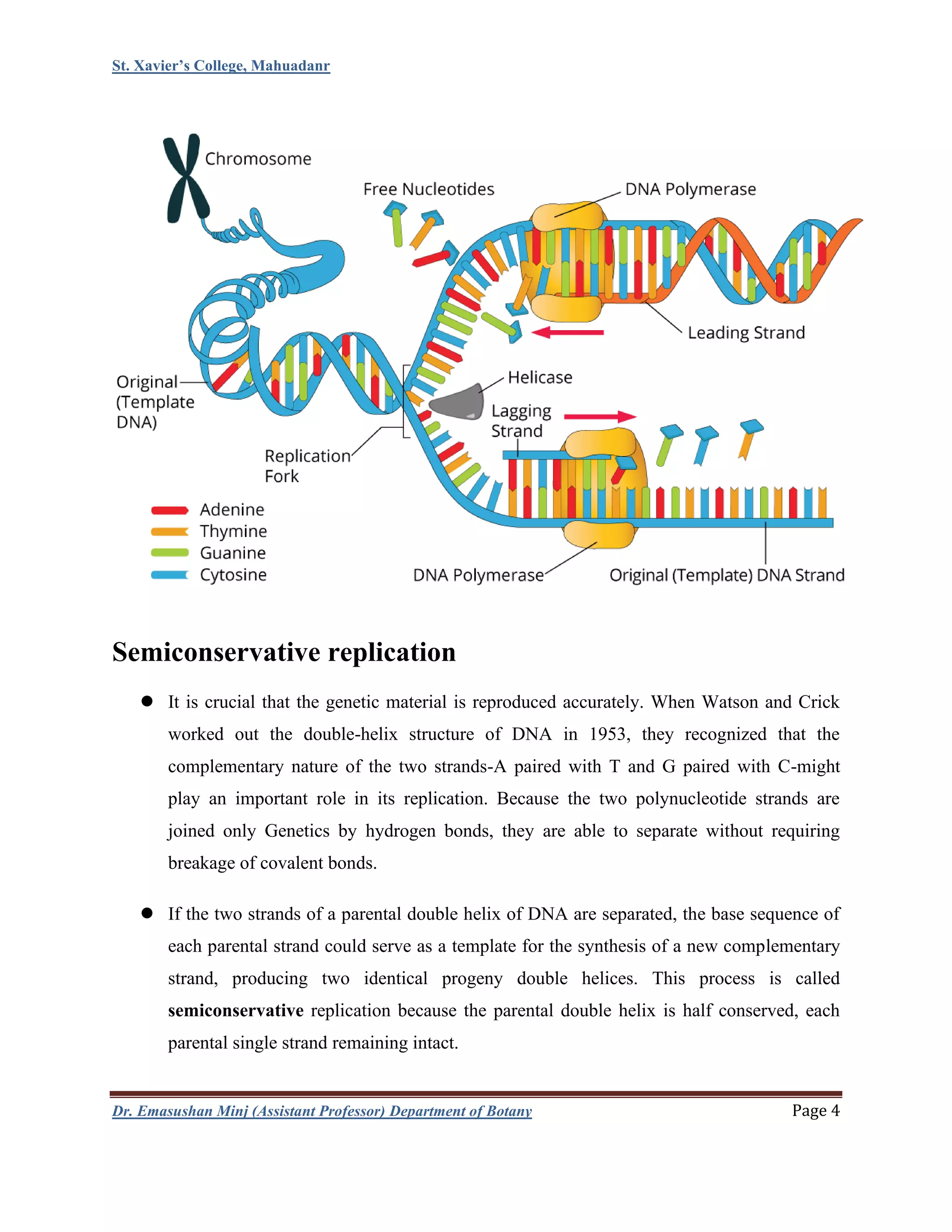
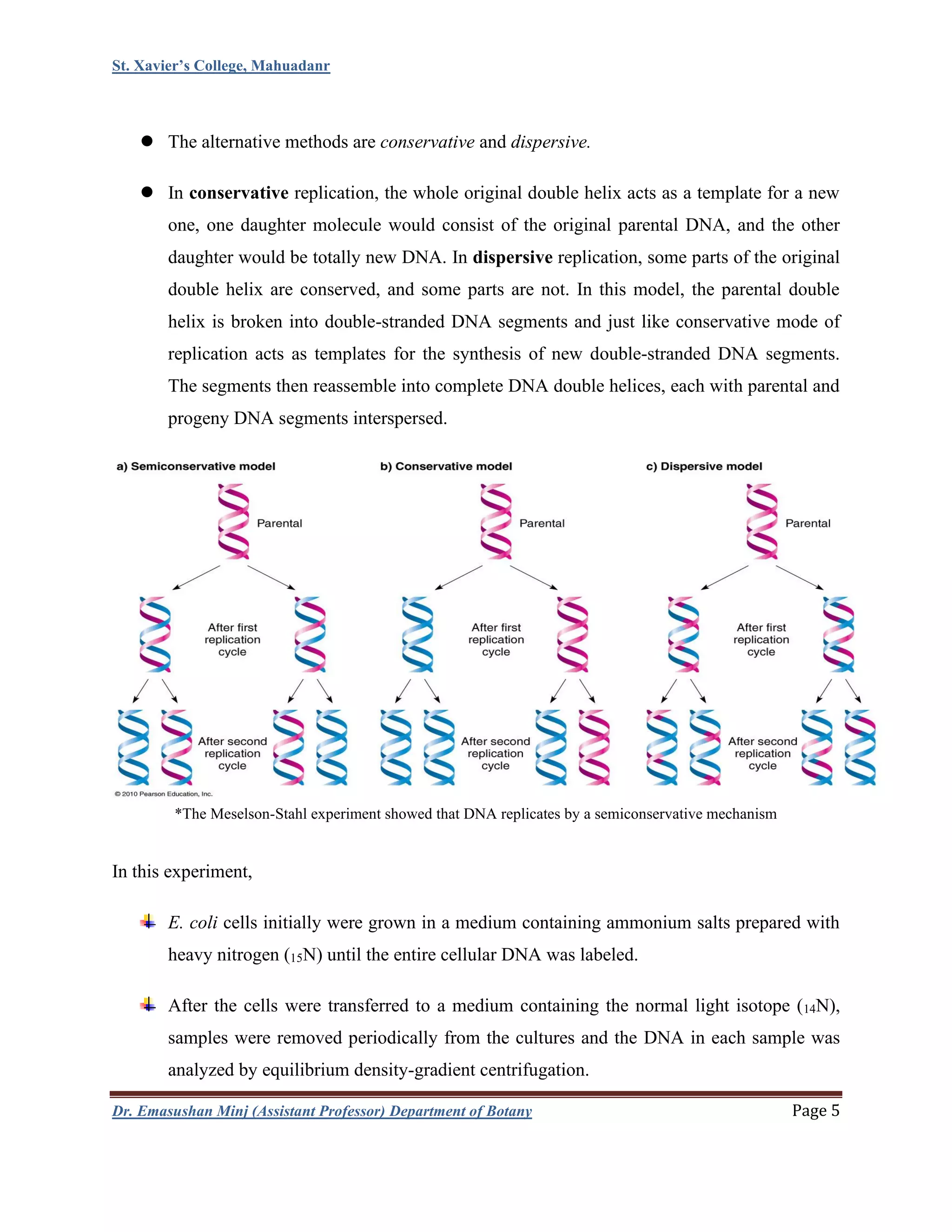
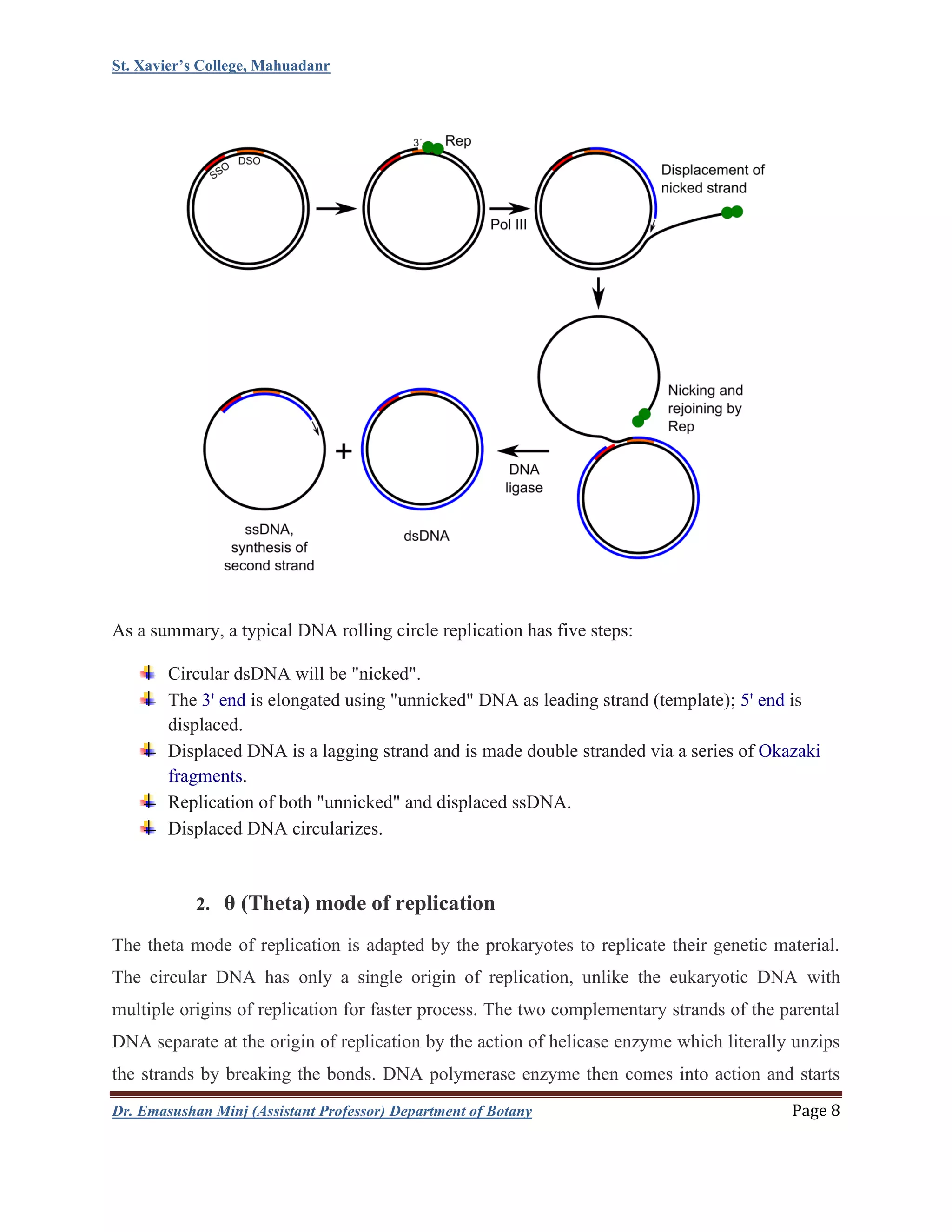
This document summarizes DNA replication in eukaryotic cells. It begins with an overview of DNA replication, including that it occurs during S phase and produces two identical DNA molecules from one original. It then describes the process of initiation, elongation, and termination of DNA replication. Initiation involves unwinding of DNA and formation of replication forks. Elongation involves continuous synthesis of the leading strand and discontinuous synthesis of the Okazaki fragments on the lagging strand. The document discusses several models of replication, including rolling circle replication, theta replication in prokaryotes, and replication of linear and telomeric DNA. It highlights key aspects like semiconservative replication being shown by Meselson-Stahl experiments. In