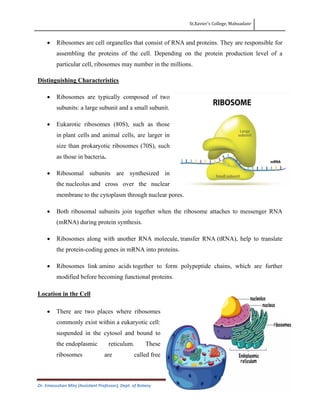
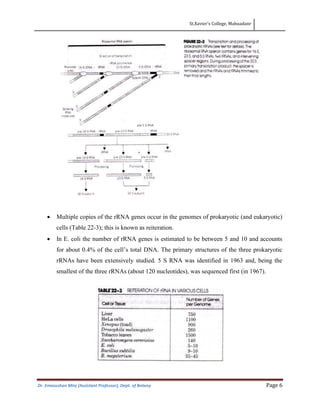
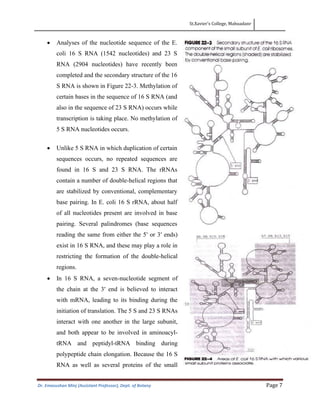
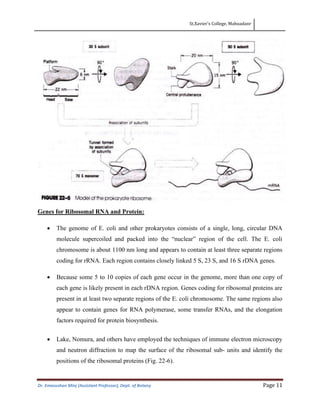
This document discusses the structure, composition, and assembly of ribosomes. It provides details on both prokaryotic and eukaryotic ribosomes. Some key points include:
- Ribosomes are composed of RNA and proteins and are found in the cytoplasm and organelles of cells. They are responsible for protein synthesis.
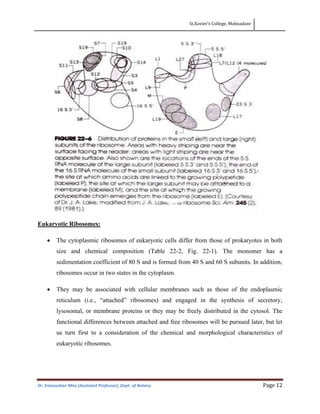
- Prokaryotic ribosomes are typically 70S and composed of 30S and 50S subunits, while eukaryotic ribosomes are larger at 80S and composed of 40S and 60S subunits.
- Ribosomes translate mRNA into polypeptide chains with the help of tRNA and link amino acids together to form proteins.