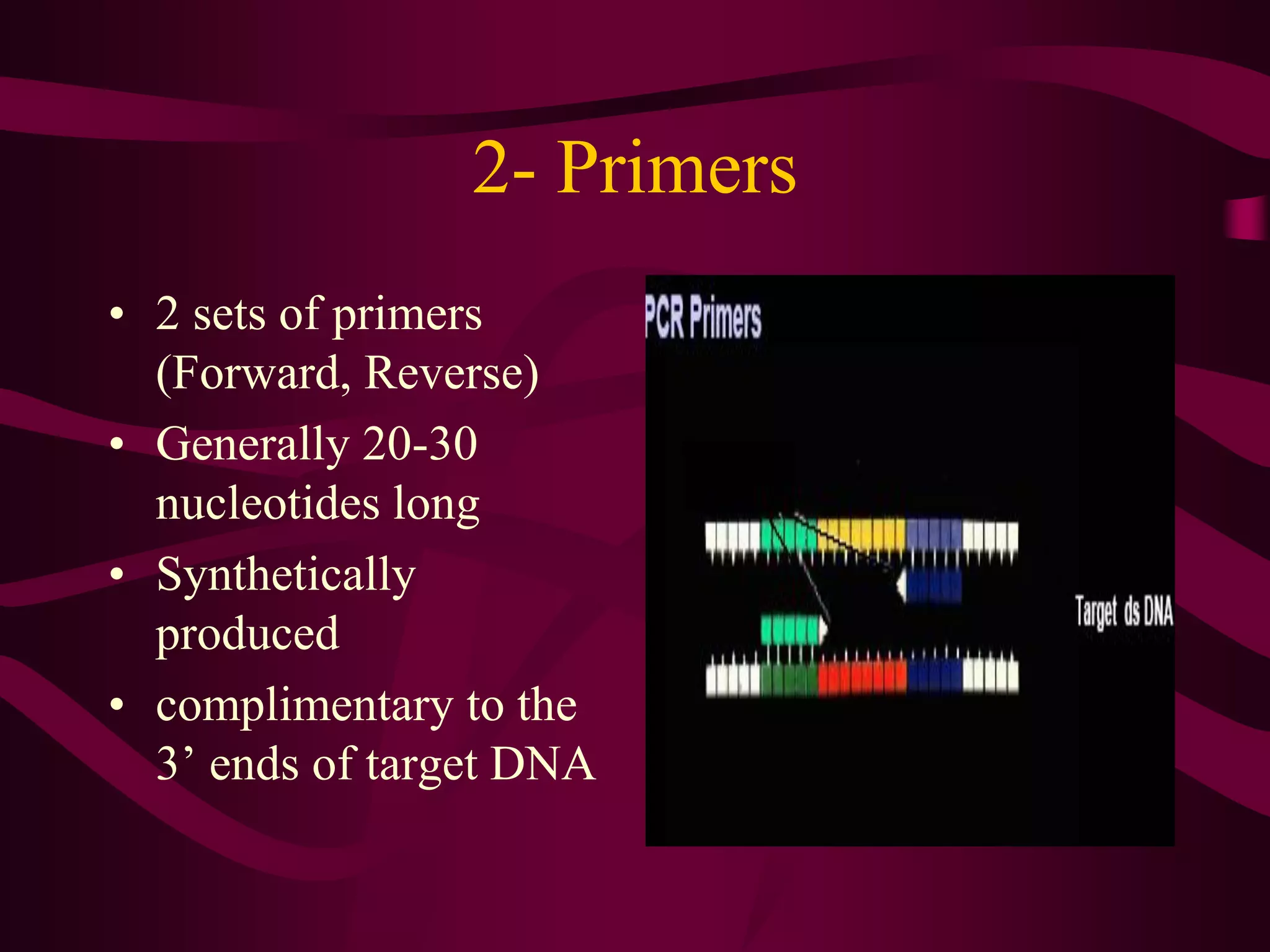
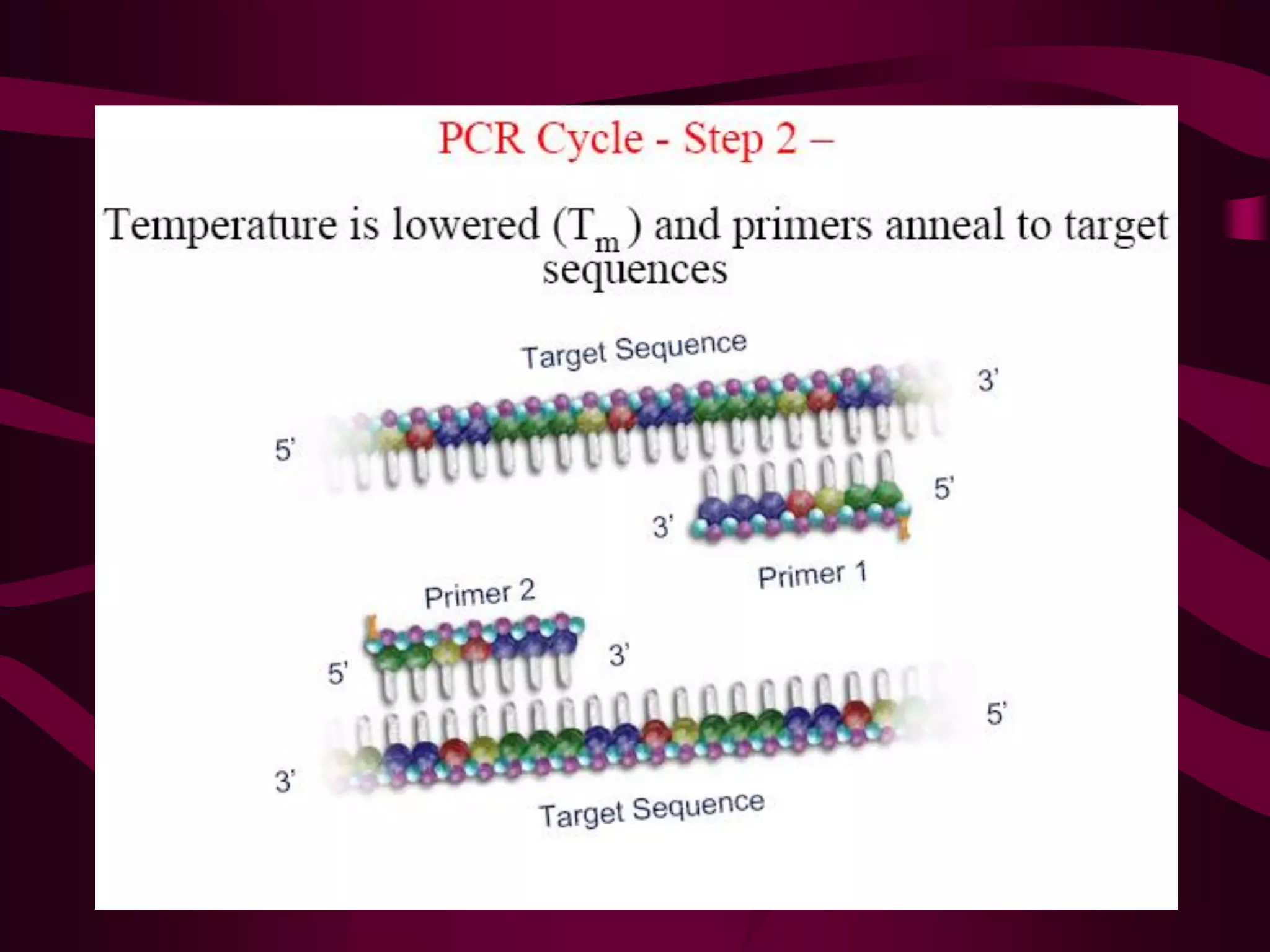
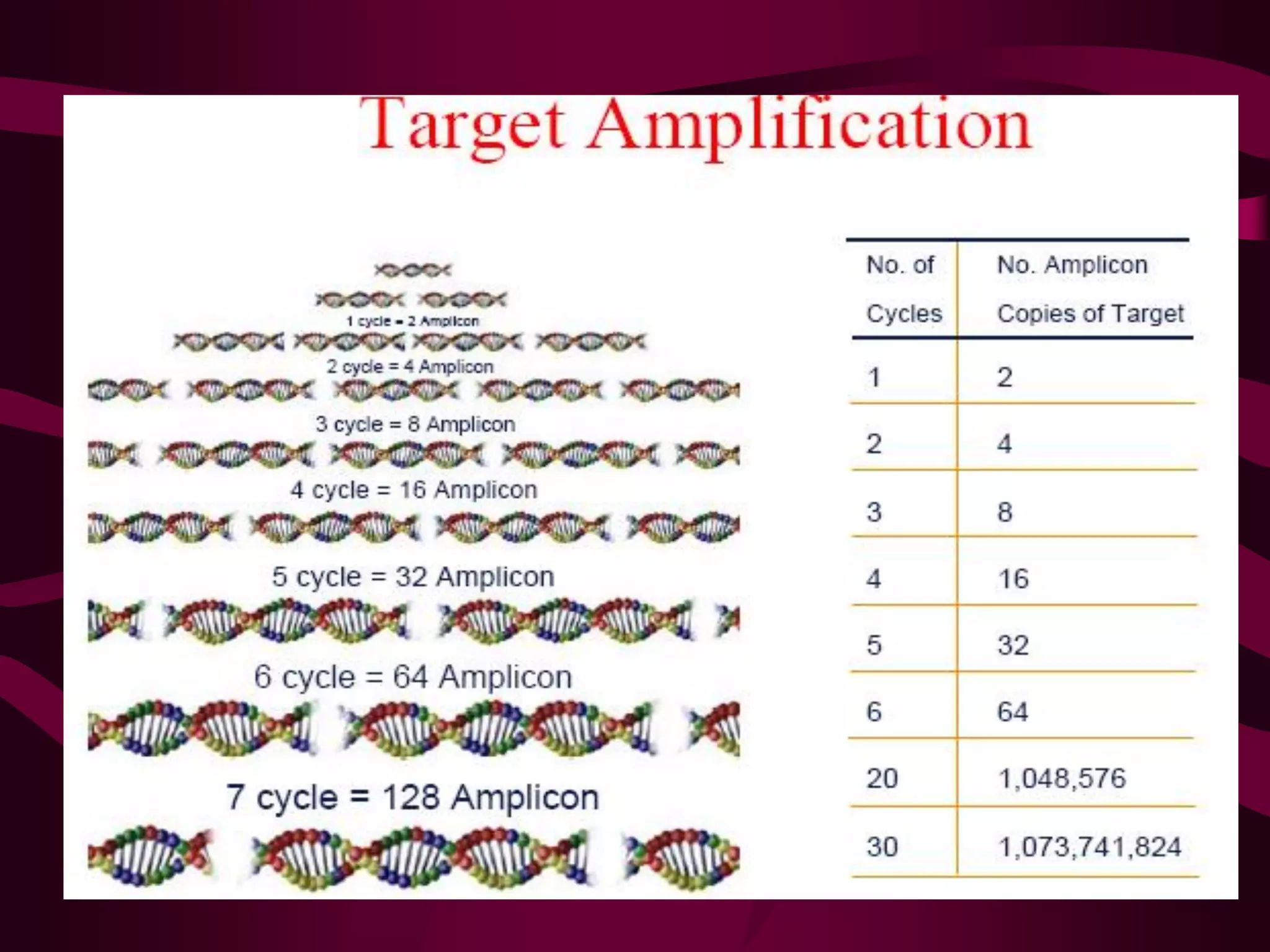
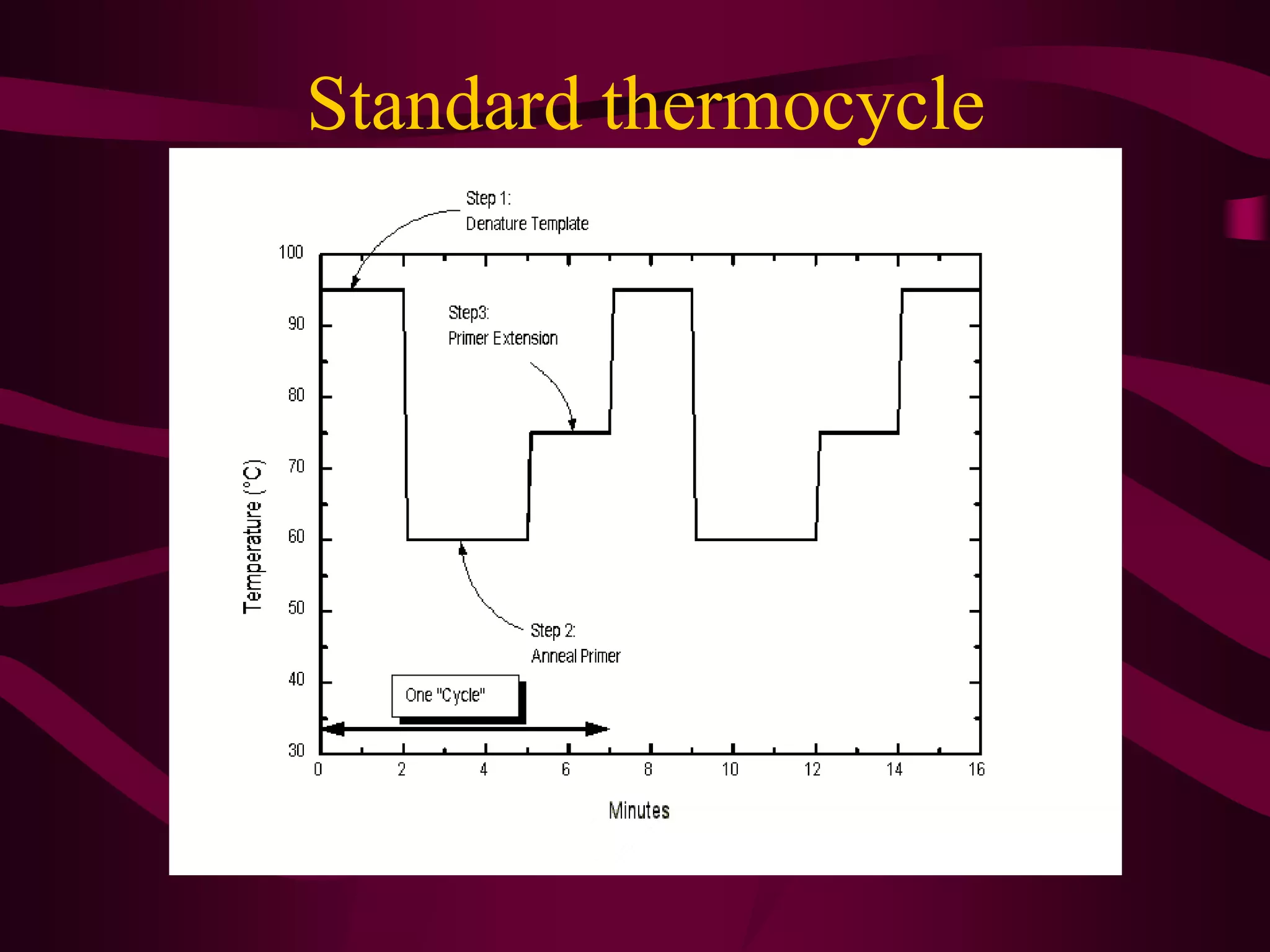
PCR is a laboratory technique used to amplify a specific region of DNA through multiple cycles of heating and cooling. It allows a small amount of DNA to be exponentially amplified into millions of copies. The key components of PCR are DNA template, primers, DNA polymerase, and nucleotides. During each cycle, the DNA is denatured, primers anneal to the DNA, and the polymerase extends the primers to synthesize new DNA strands. This allows the specific target sequence to be rapidly amplified. There are various types of PCR that have different applications in research, forensics, and medicine.