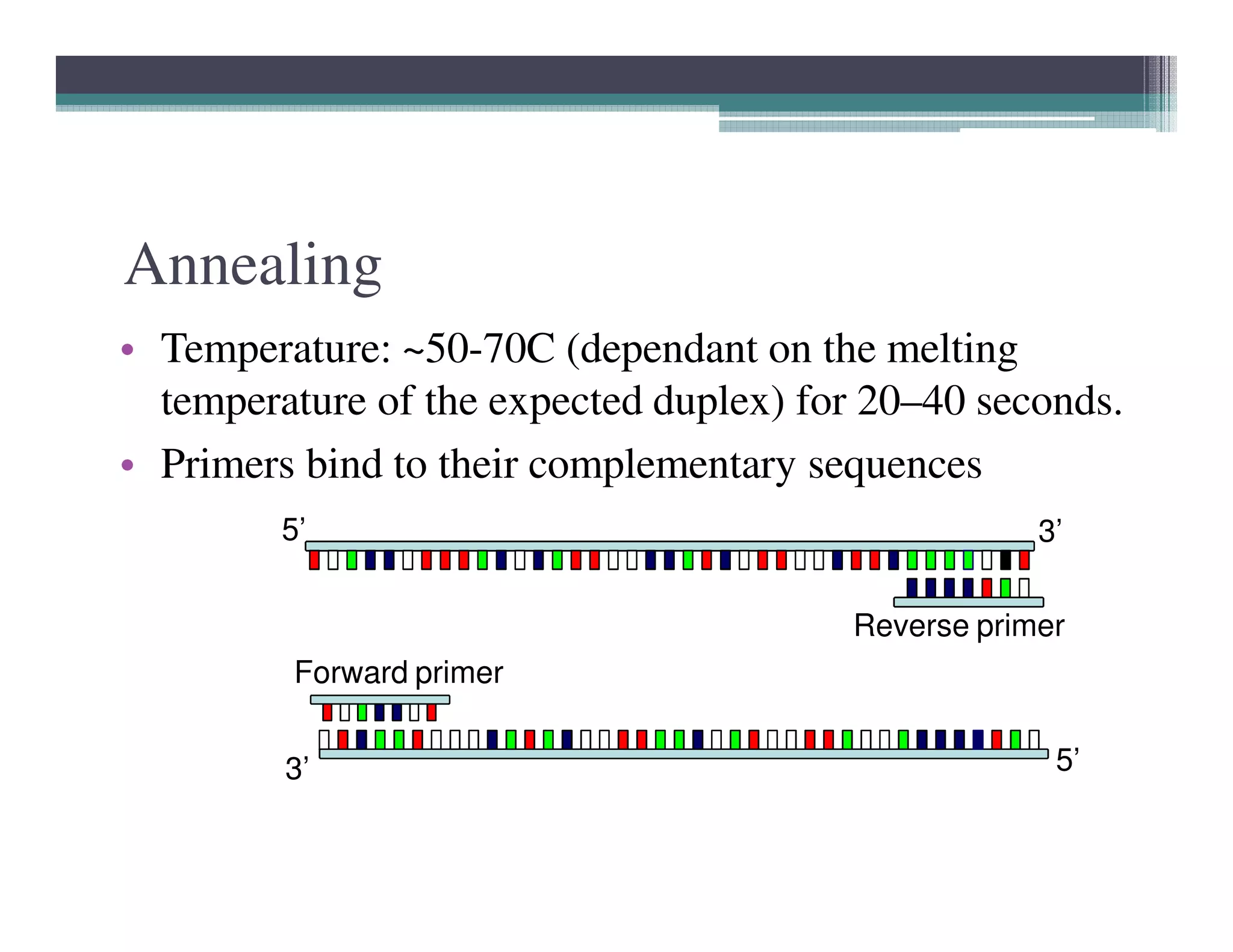
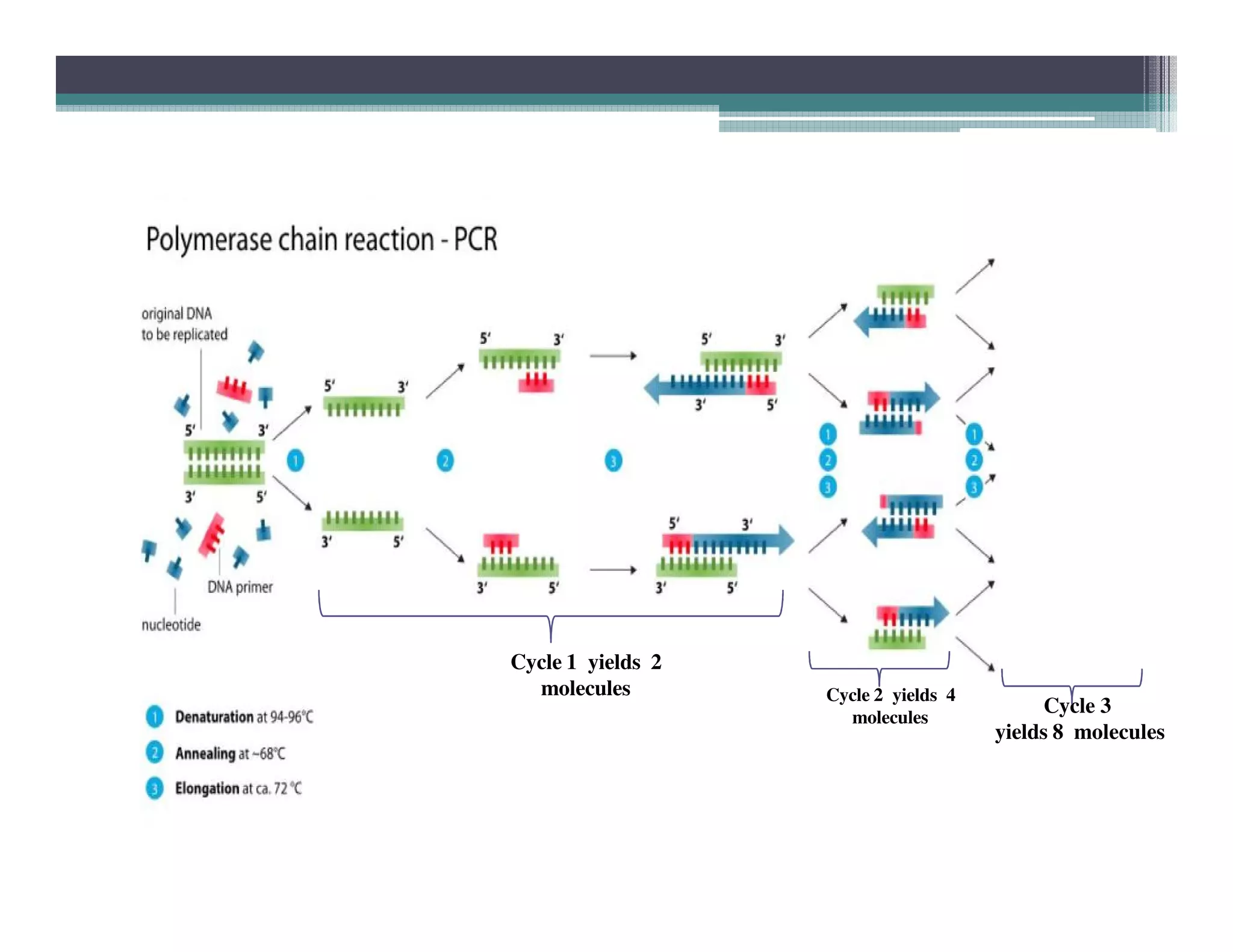
Polymerase chain reaction (PCR) is a technique used to amplify a single copy of a DNA segment across orders of magnitude, generating thousands to millions of copies. It was invented in 1984 by Kary Mullis and is now commonly used in clinical and research applications. PCR uses DNA polymerase to amplify a target DNA segment defined by primer sequences. It involves repeated cycles of heating and cooling of the DNA sample to separate, anneal, and extend the DNA strands. Mullis received the Nobel Prize in Chemistry in 1993 for his work inventing PCR.