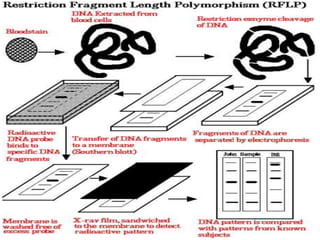
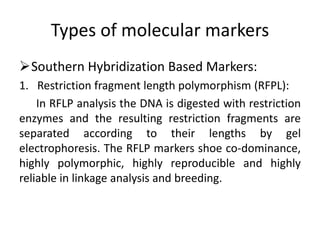
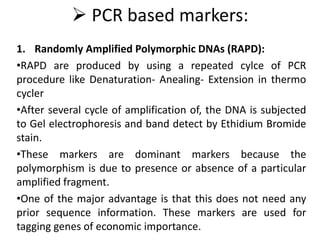
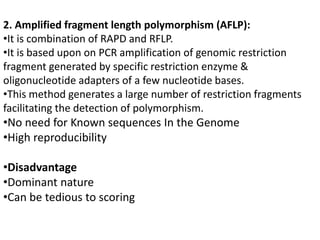
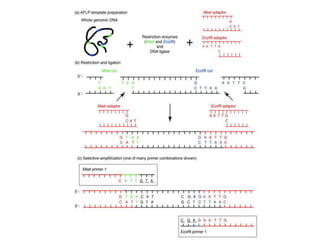
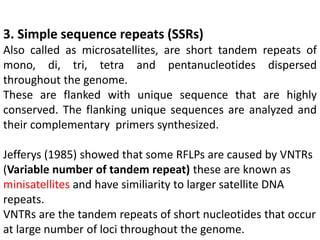
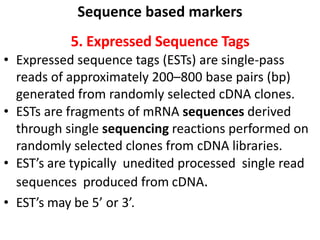
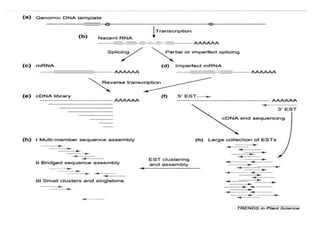
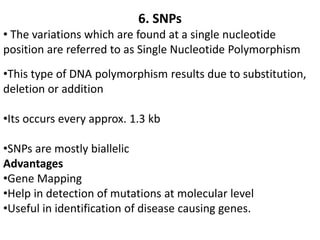
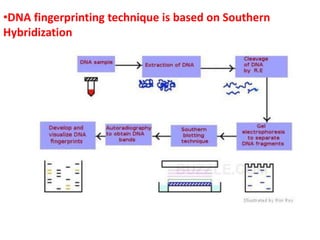
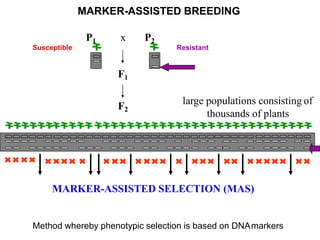
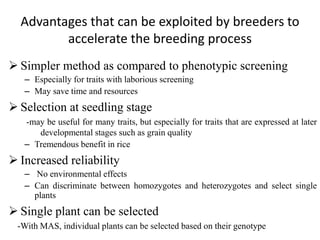
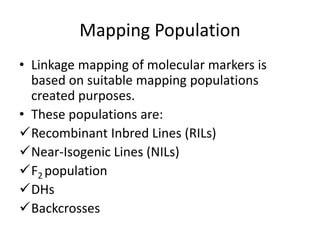
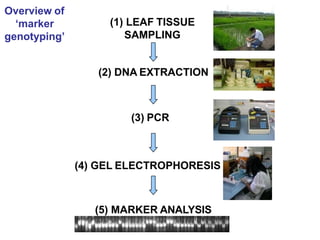
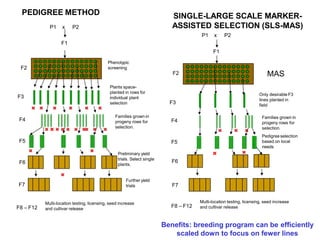
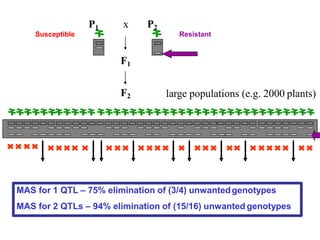
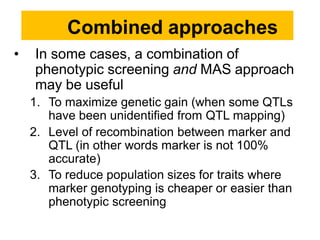
The document provides a comprehensive overview of molecular markers and marker-assisted selection in agricultural breeding, defining markers as genetic elements that aid in identifying desirable traits. It describes various types of markers, including morphological, biochemical, and DNA-based, highlighting their advantages and disadvantages for plant breeding. Additionally, it illustrates the marker-assisted selection process as a means to improve crop varieties more efficiently by utilizing genetic information.