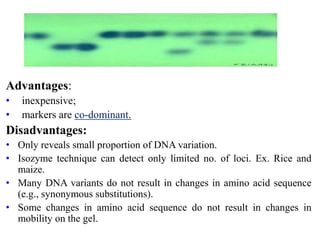
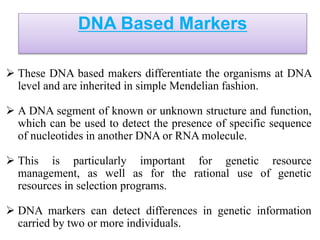
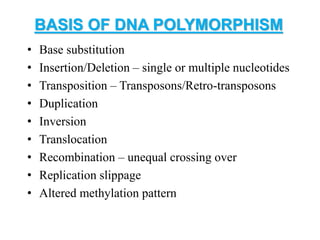
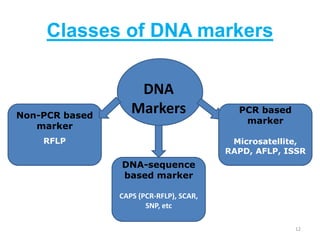
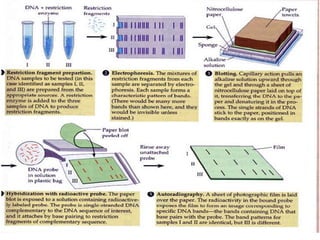
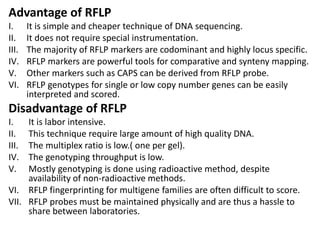
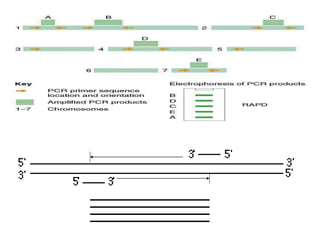
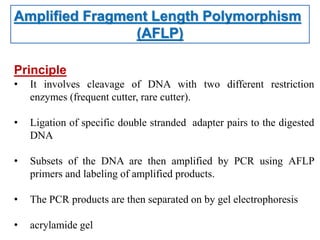
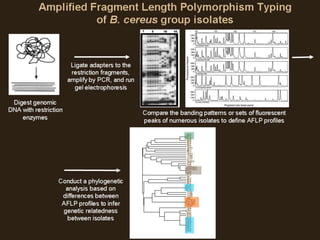
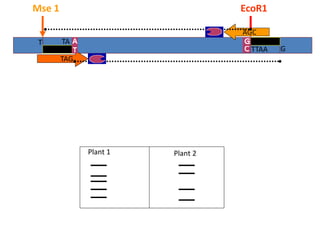
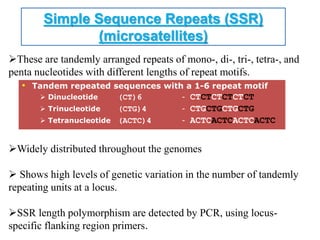
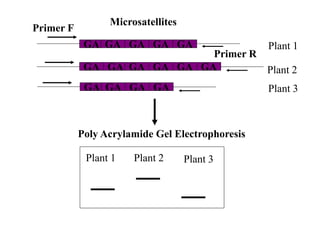
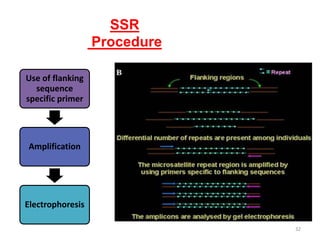
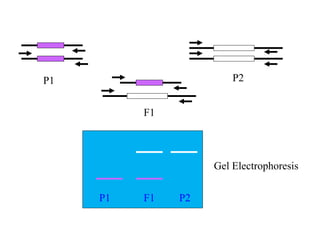
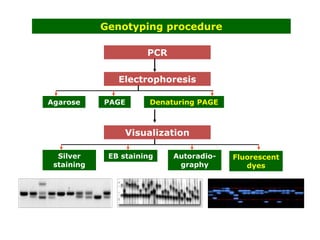
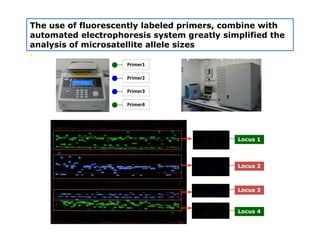
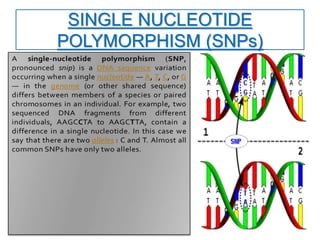
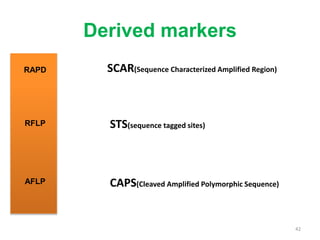
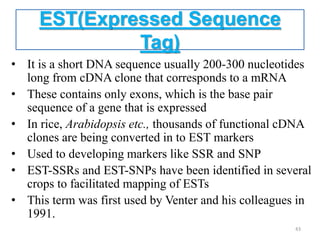
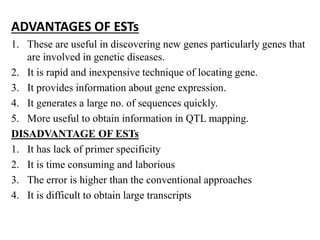
Molecular markers such as RFLP, RAPD, AFLP, SSR, SNPs, and ESTs can be used to detect polymorphisms at the DNA level. SSR markers, also known as microsatellites, detect length polymorphisms in tandem repeats of short nucleotide motifs. SSRs are widely distributed in genomes and show high levels of variation, making them useful for applications like genetic mapping, variety identification, and marker-assisted selection.