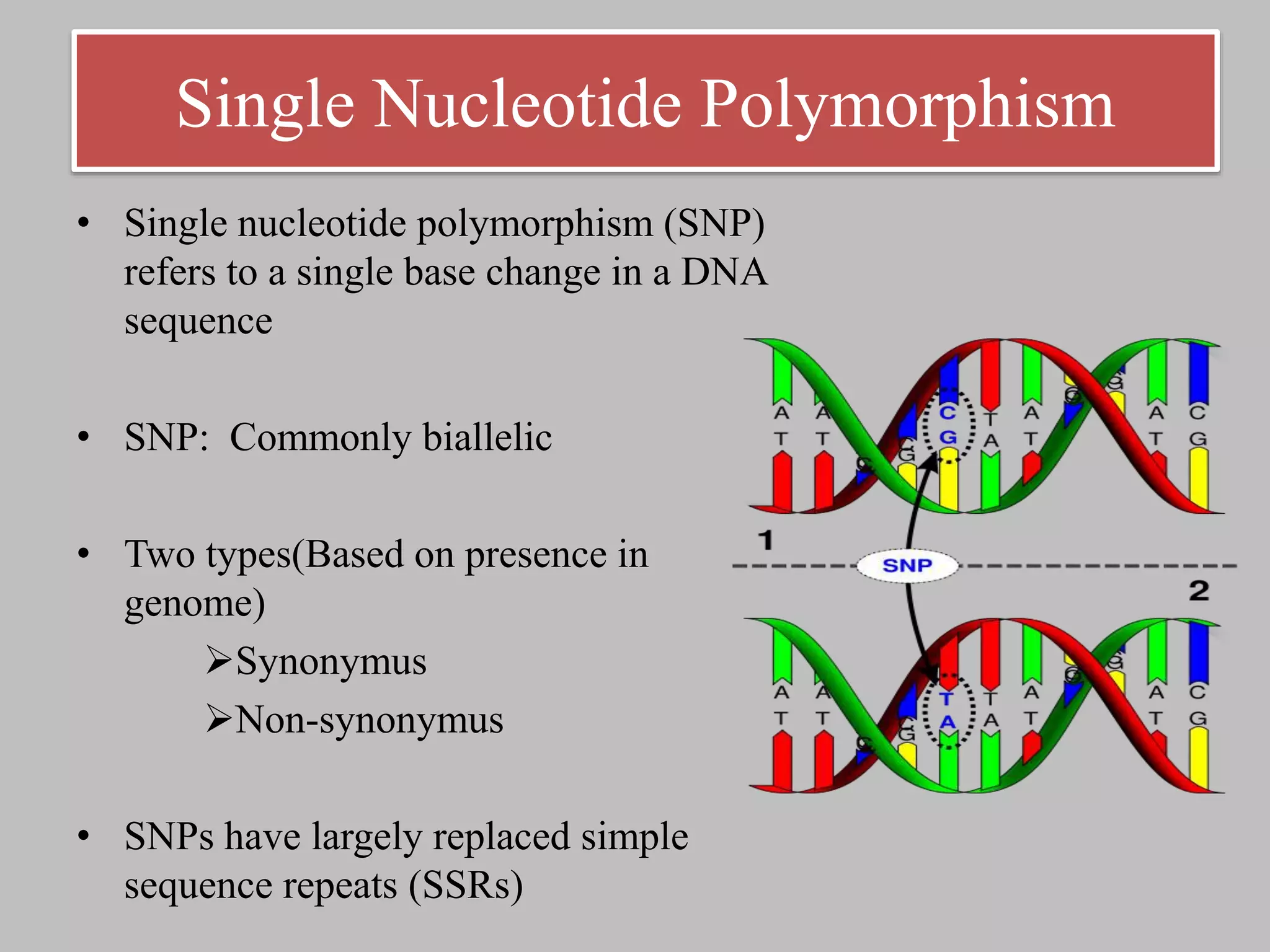
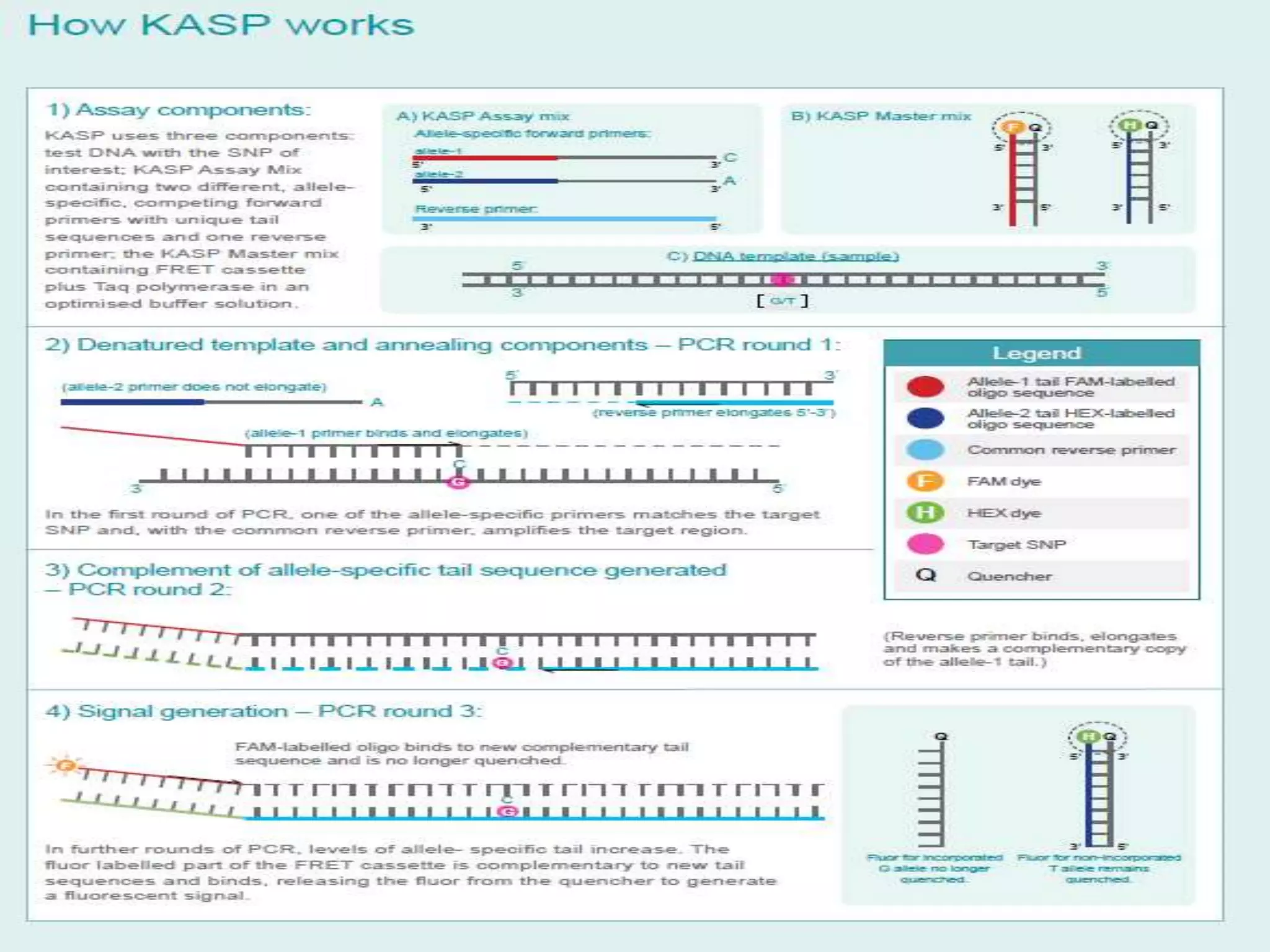
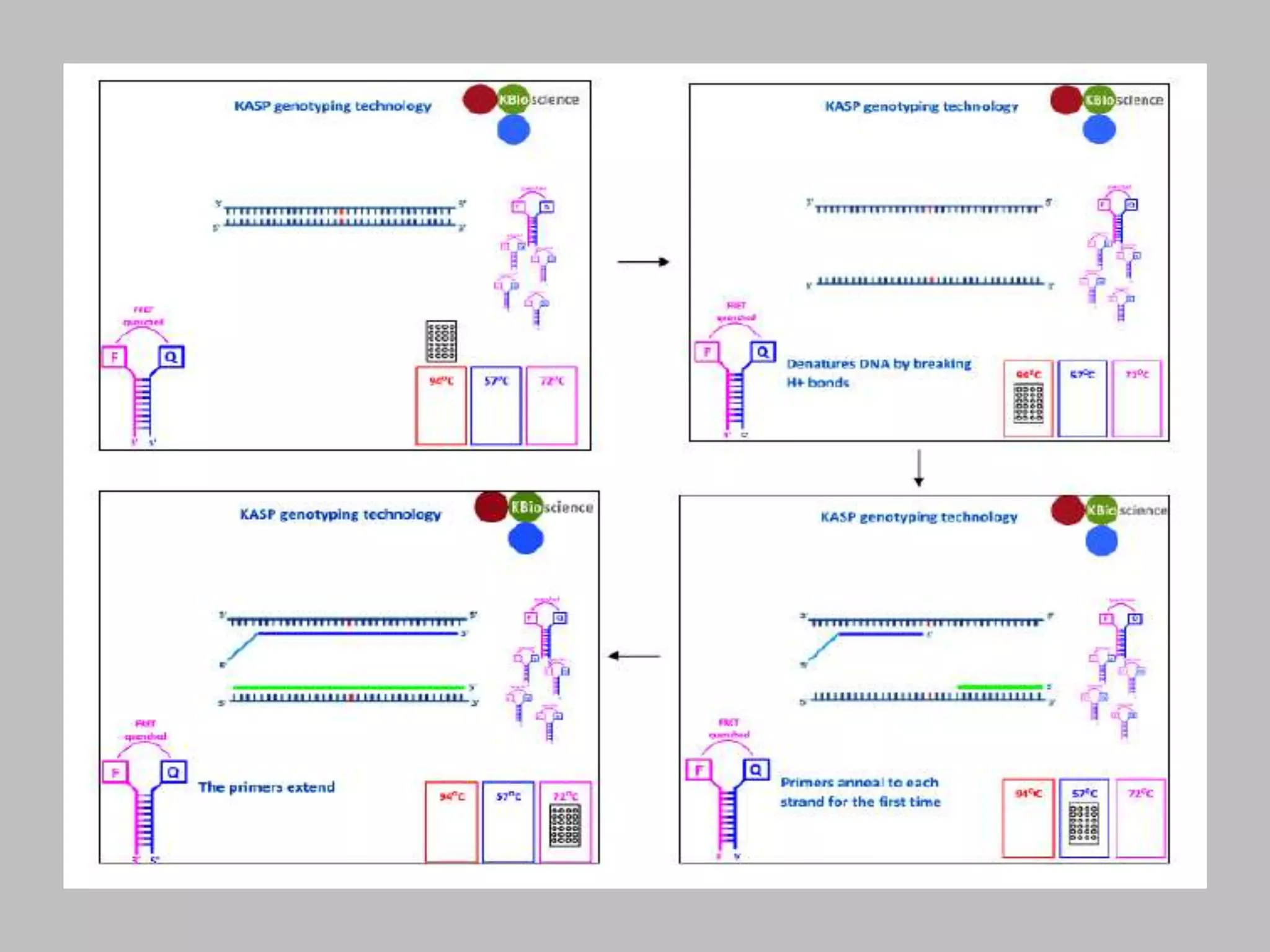
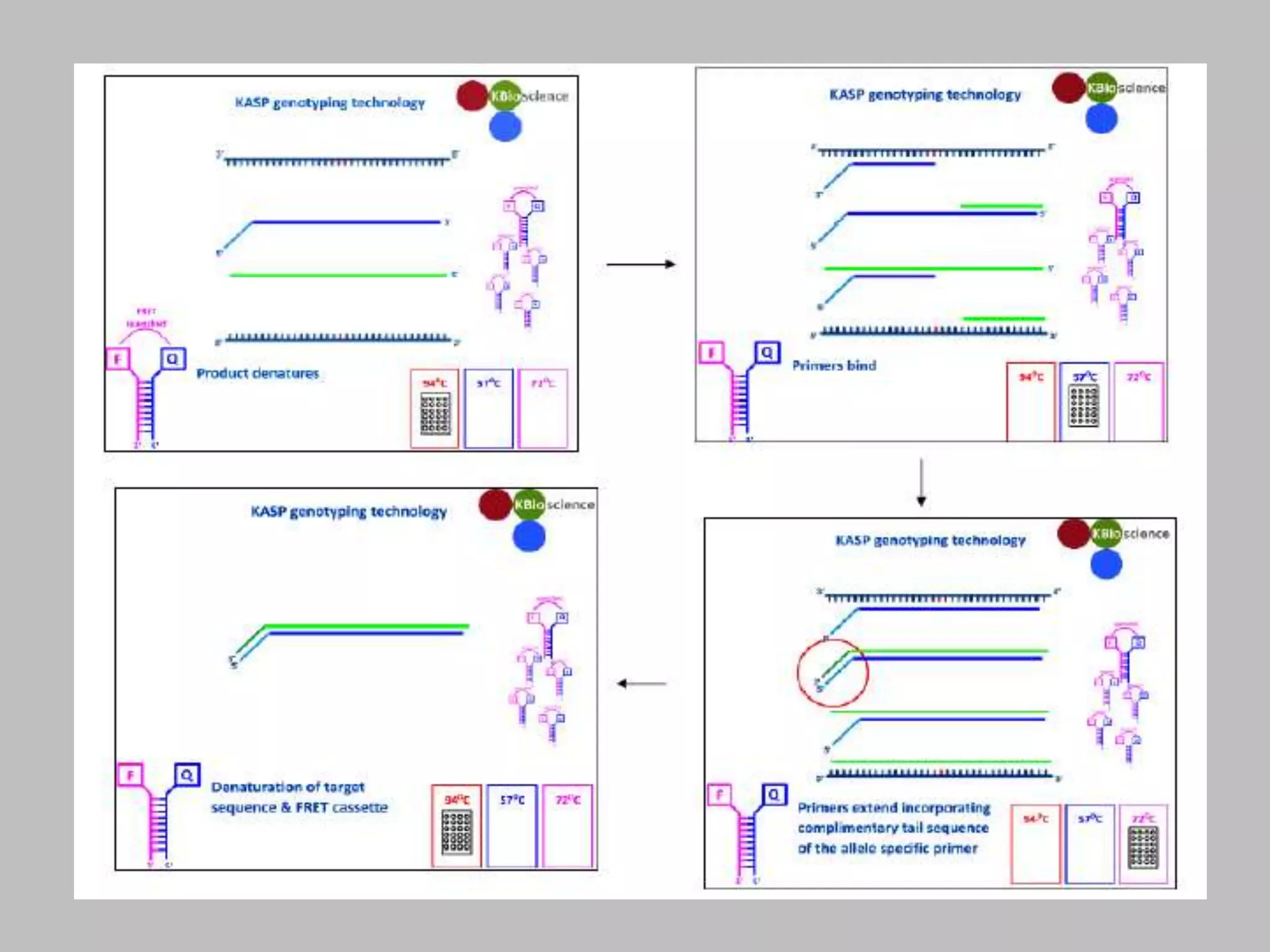
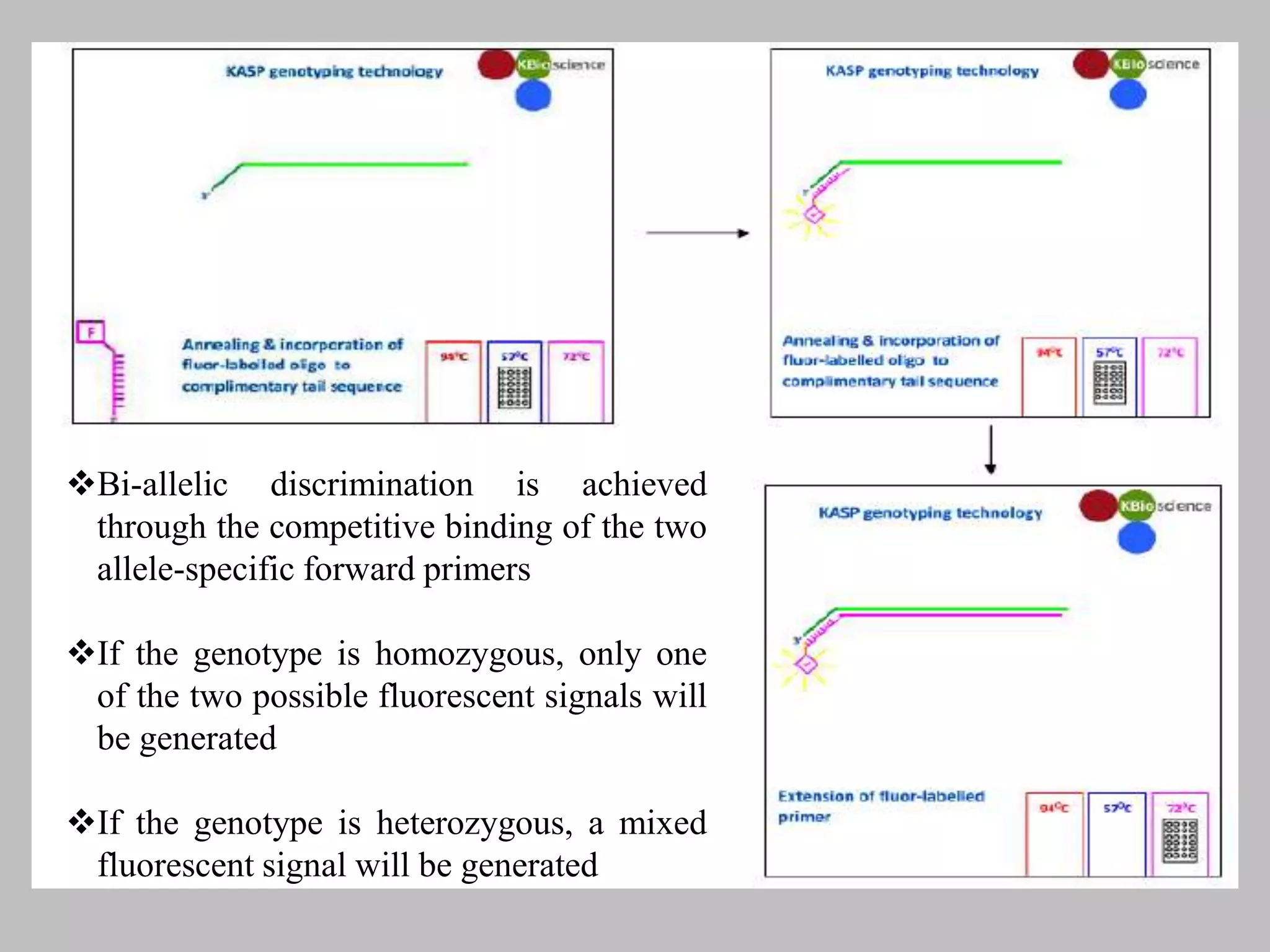
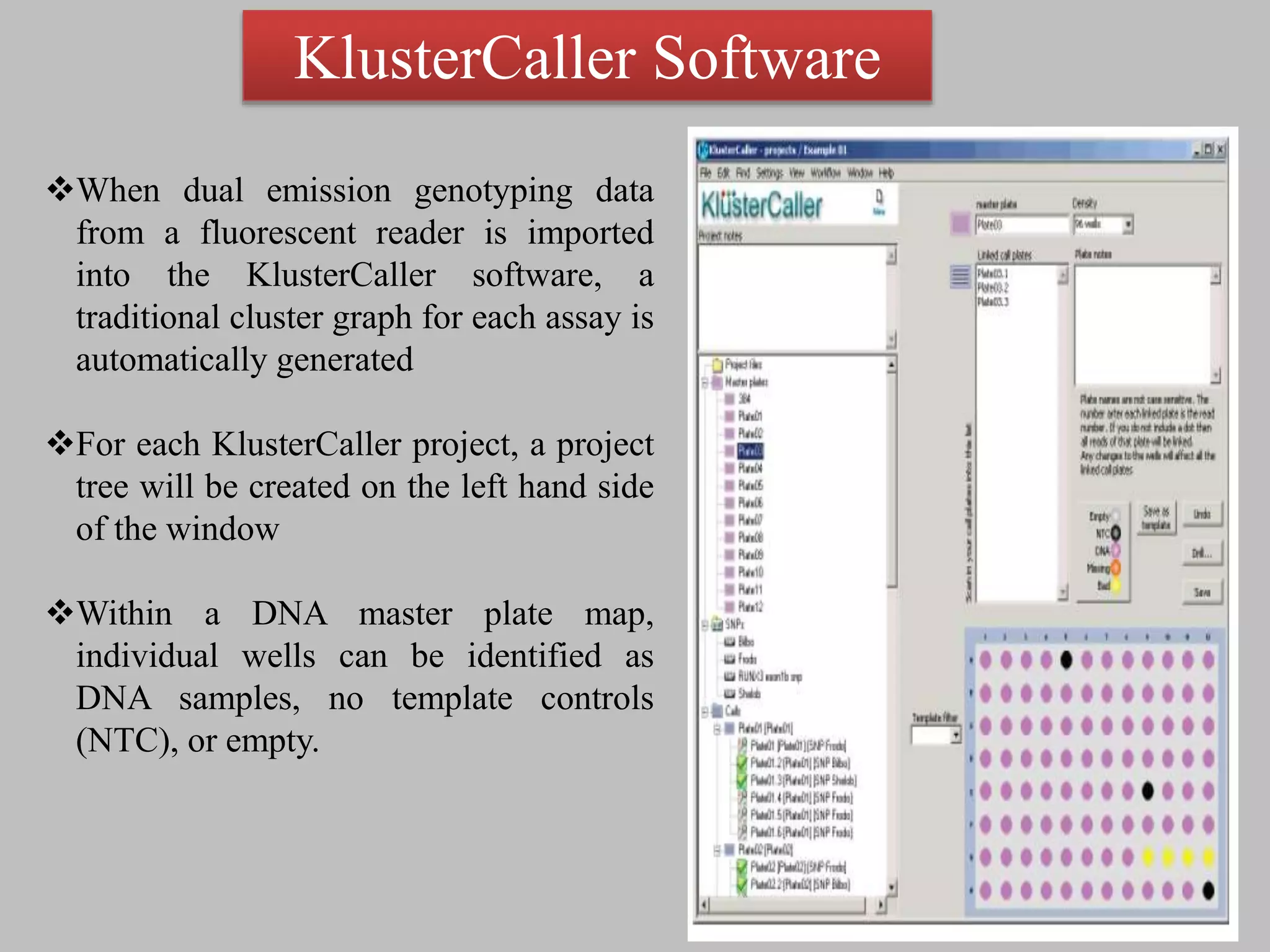
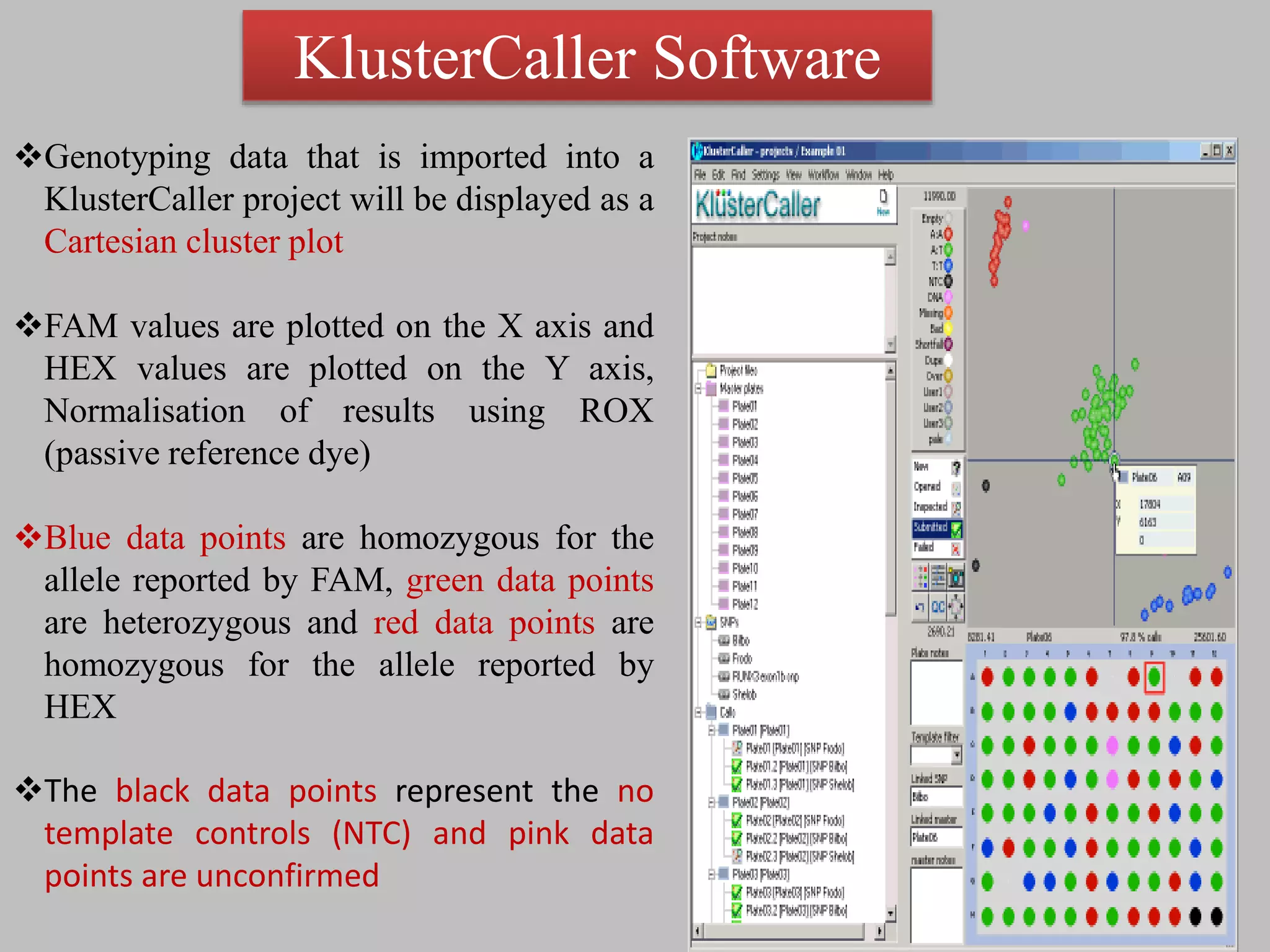
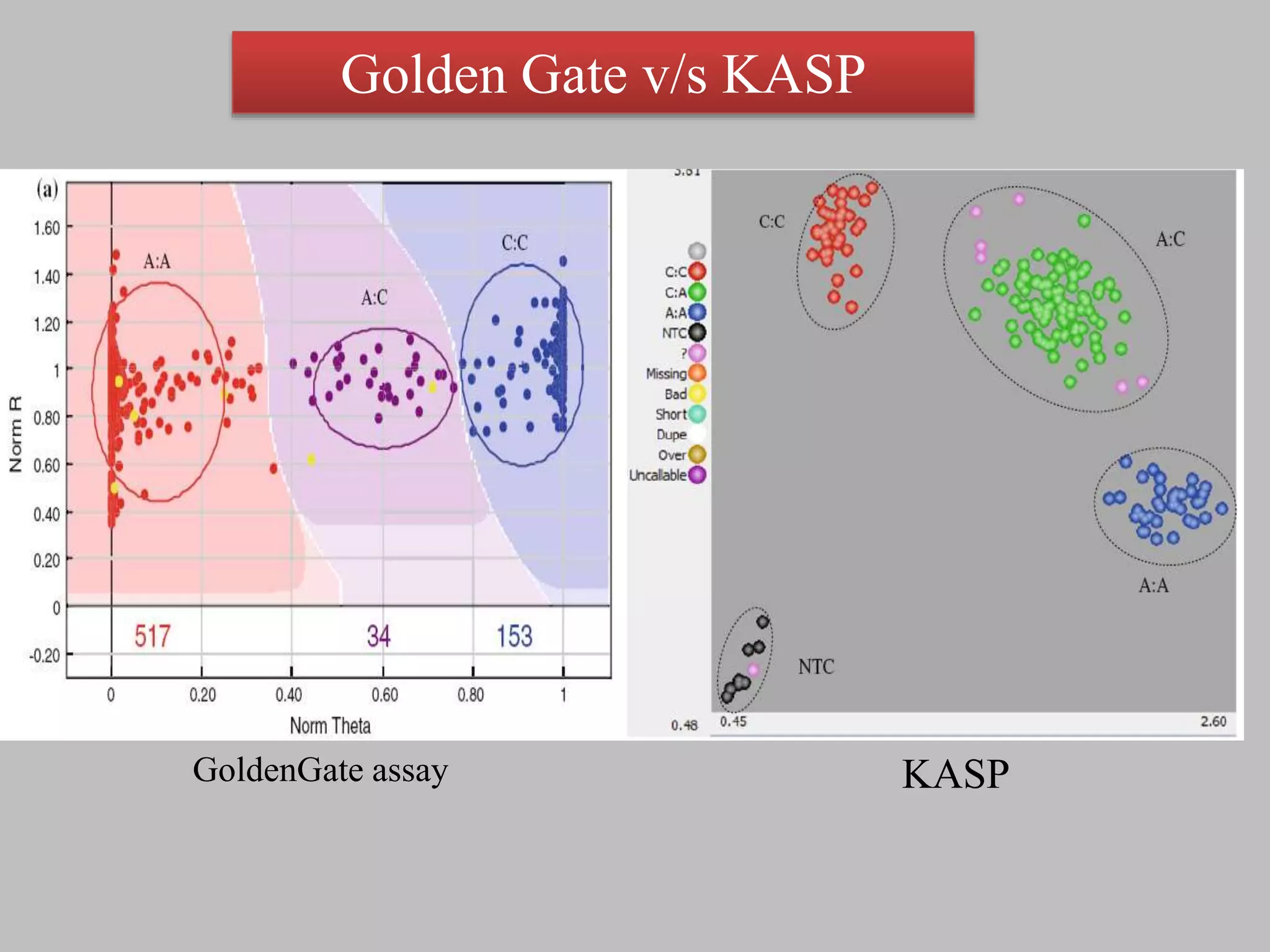
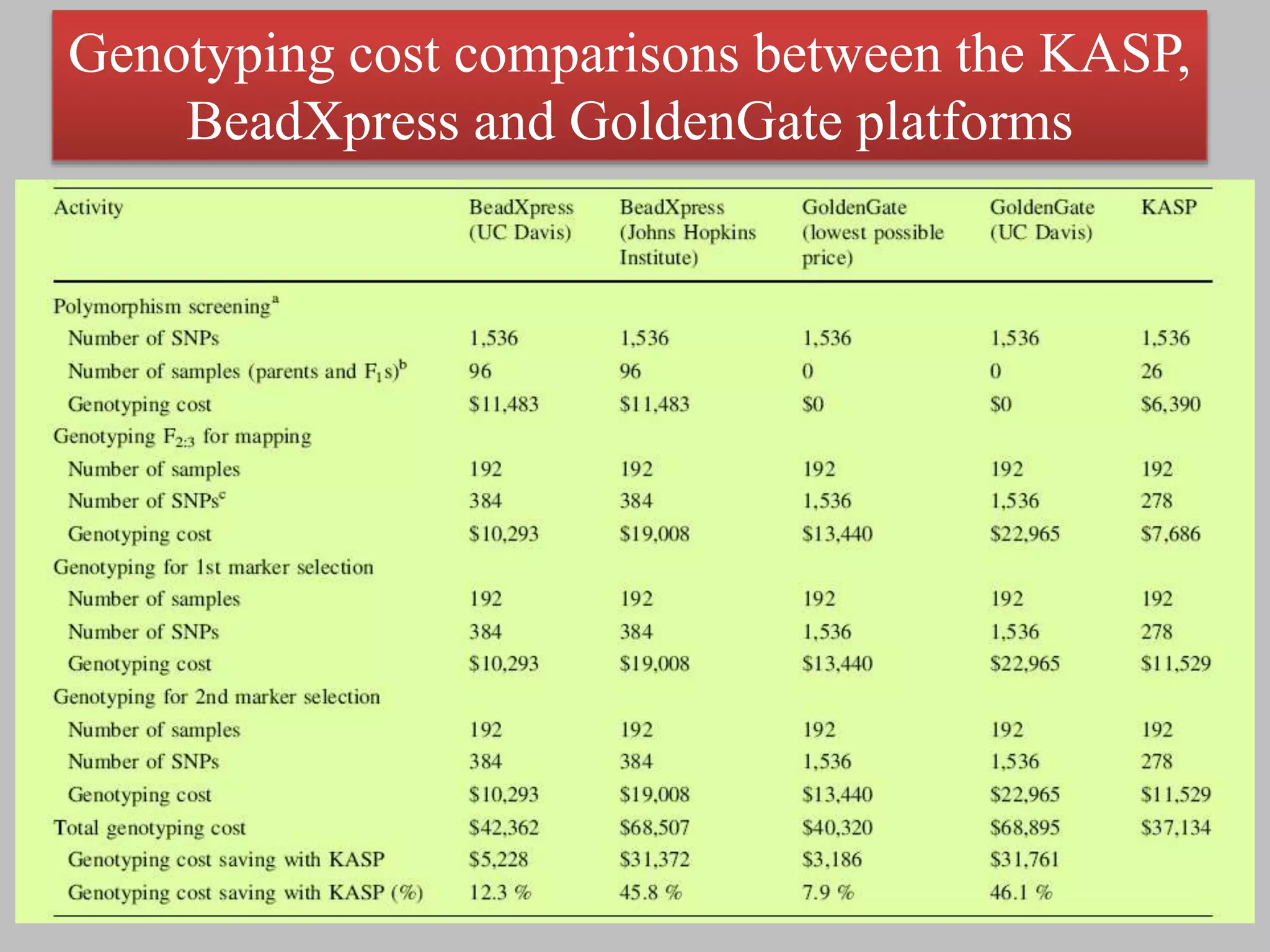
The document discusses single nucleotide polymorphism (SNP) genotyping using Kompetitive Allele Specific PCR (KASP), highlighting its advantages such as low assay cost, high genomic abundance, and co-dominant inheritance. It details the KASP technology, its components, the importance of quality control, and applications in areas like quantitative trait loci (QTL) mapping and allele mining. Additionally, various SNP genotyping platforms are compared, alongside data handling with Klustercaller software for efficient analysis.