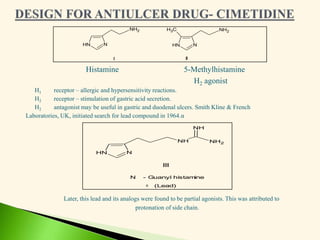
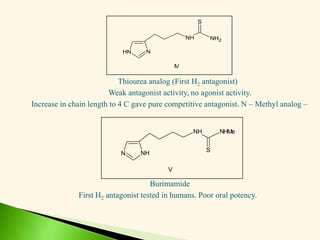
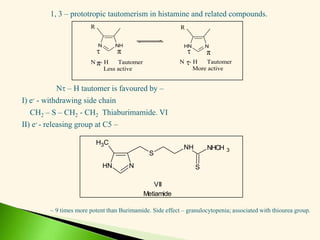
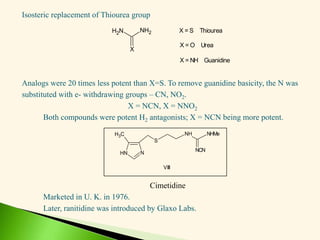
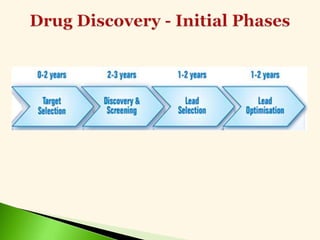
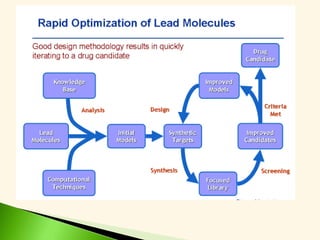
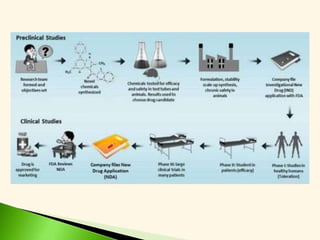
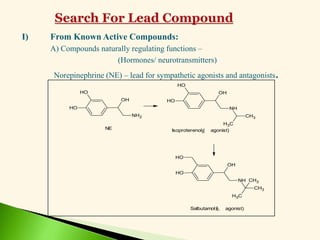
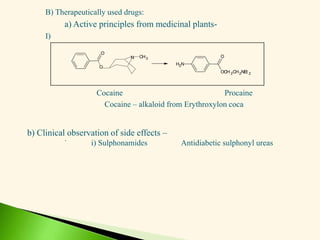
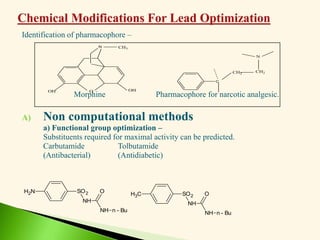
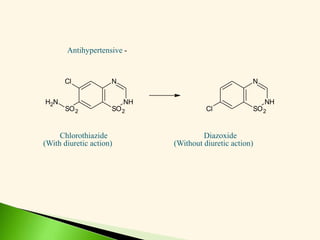
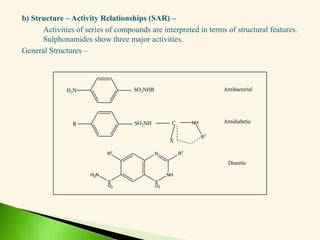
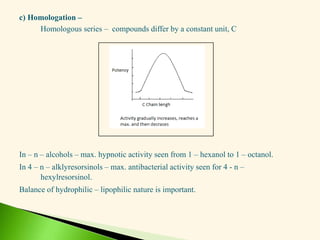
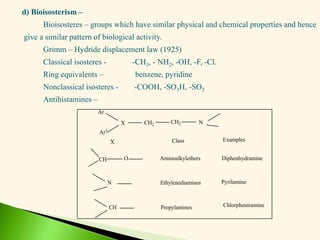
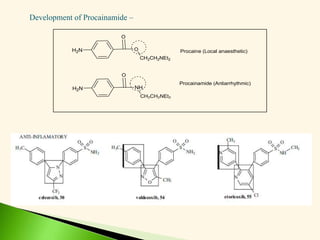
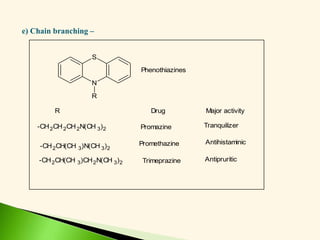
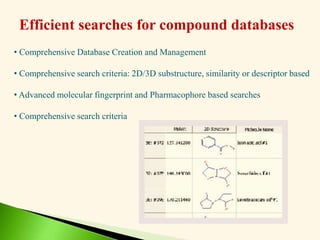
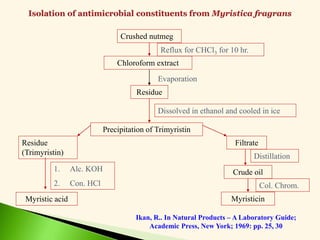
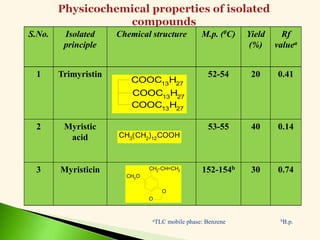
The document discusses the drug discovery process, emphasizing lead compounds and various methodologies for identifying and modifying them for therapeutic use. It covers concepts such as bioassays, molecular manipulation, structure-activity relationships, and quantitative structure-activity relationships in drug development, including examples from known drugs and natural products. Additionally, it details synthesis methods, isolation of active principles, and the application of computational techniques for predicting biological activity.










![Scheme for syntheses of myristic acid derivatives
CH3
[CH2
]12
COOH
ROH/H2
SO4
[M1]
[M2 - M9, M21- M23]
SOCl2
Amine
[M11 - M20, M24 - M27]
CH3[CH2]12COOR
CH3[CH2]12COOH
[M1]
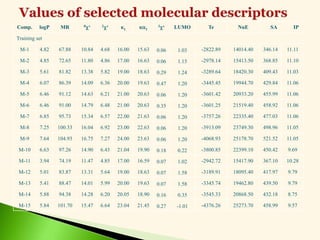
CH3
[CH2
]12
COCl
CH3[CH2]12COR
ROH
CH3
[CH2
]12
COOR
[M10]](https://image.slidesharecdn.com/leadoptimization-220705030809-3a3322ea/85/Lead-Optimization-in-Drug-Discovery-32-320.jpg)

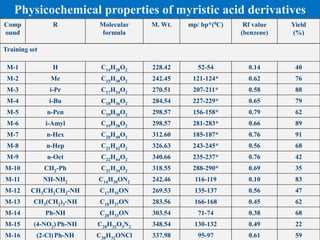
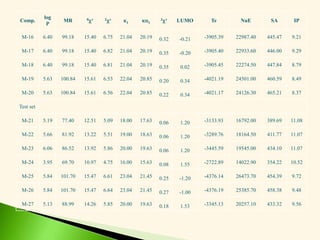
![Compou
nd
R Molecular
formula
M. Wt. mp/ bp*(0C) Rf value
(benzene)
Yield
(%)
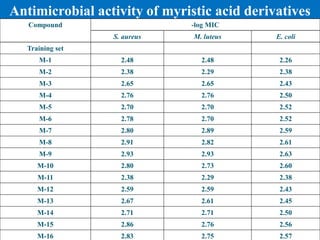
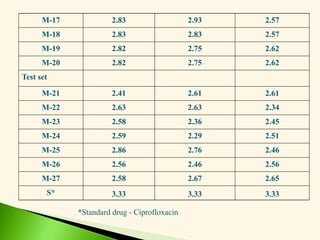
M-17 (3-Cl) Ph-NH C20H32ONCl 337.98 136-138 0.42 69
M-18 (4-Cl) Ph-NH C20H32ONCl 337.98 115-117 0.54 72
M-19 (2-CH3O) Ph-
NH
C21H35O2N 333.57 156-158 0.67 86
M-20 (4-CH3O) Ph-
NH
C21H35O2N 333.57 165-167 0.58 46
Test set
M-21 Et C16H32O2 256.48 180-182* 0.60 82
M-22 n-Pr C17H34O2 270.51 217-219* 0.61 66
M-23 n-Bu C18H36O2 284.54 271-273* 0.58 74
M-24 NH2 C14H29ON 227.44 80-82 0.10 87
M-25 (2-NO2) Ph-NH C20H32O3N2 348.54 146-148 0.45 18
M-26 (3-NO2) Ph-NH C20H32O3N2 348.54 211-213 0.22 84
M-27 NH(Et)2 C18H37ON 283.56 68-70 0.13 24
CH3
[CH2
]12
COOR CH3
[CH2
]12
COR
[M1
- M10
, M21
- M23
] [M11
- M20
, M24
- M27
]](https://image.slidesharecdn.com/leadoptimization-220705030809-3a3322ea/85/Lead-Optimization-in-Drug-Discovery-35-320.jpg)
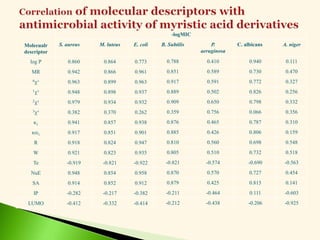

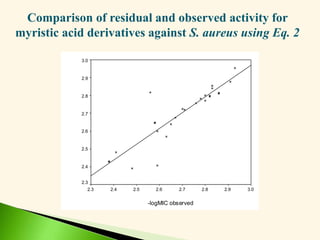
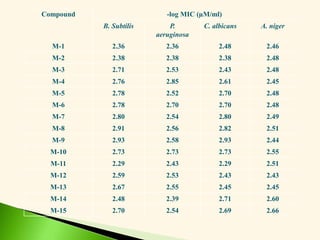
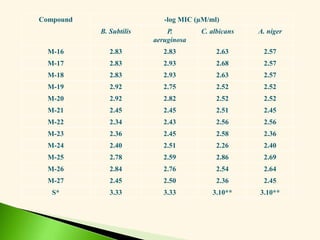

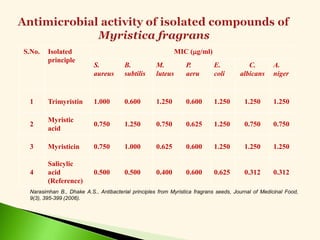
![Antibacterial activity against B. subtilis (Eq. 5 & 6)
-log MIC = 0.113 0v + 1.077 (5)
n =20 r = 0.917 F = 95.65 s = 0.079 q2 = 0.808
-log MIC = 0.244 2v + 1.187 (6)
n =20 r = 0.908 F = 85.10 s = 0.083 q2 = 0.796
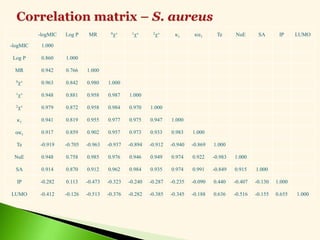
B. Narasimhan and A.S.Dhake, Theoretical modeling of antimicrobial activity of
myristic acid derivatives by Hansch analysis, National symposium on challenges in drug
discovery research: Networking opportunities between academia and Industries, Birla
Institute of Technology and Science, Pilani, April 7-8, 2006, P.93 [Abstract No. CC-902].](https://image.slidesharecdn.com/leadoptimization-220705030809-3a3322ea/85/Lead-Optimization-in-Drug-Discovery-47-320.jpg)