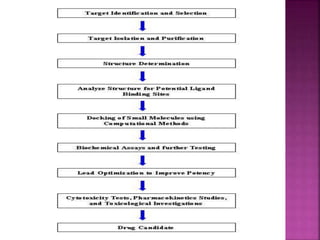
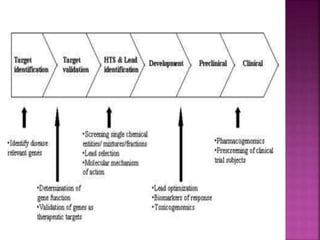
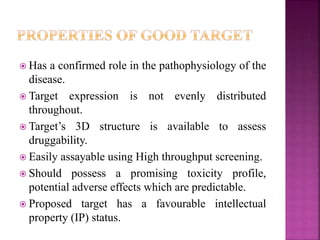
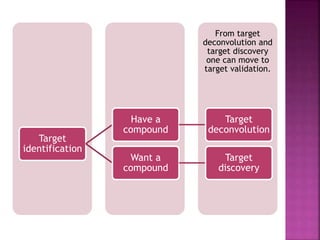
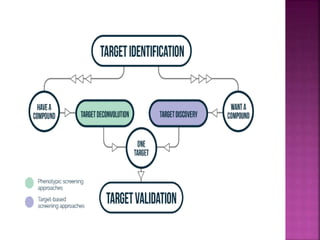
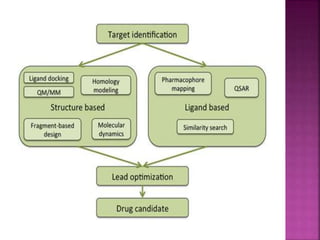
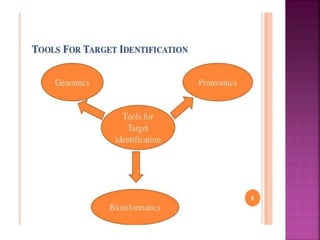
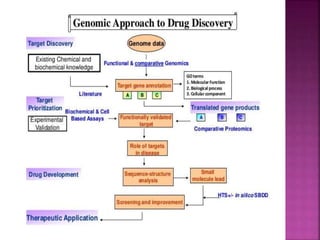
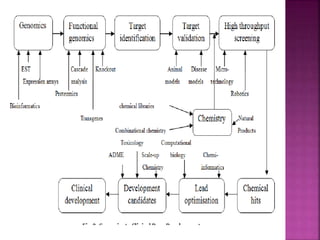
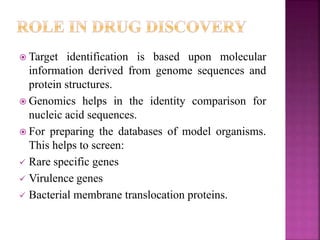
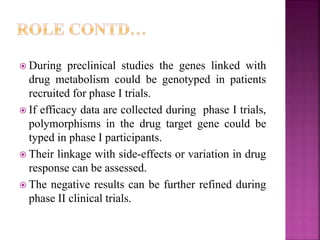
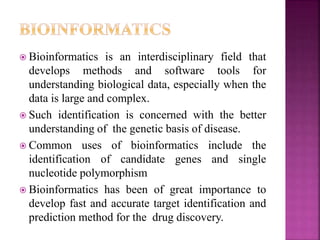
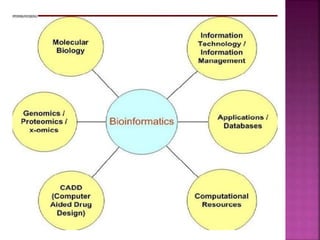
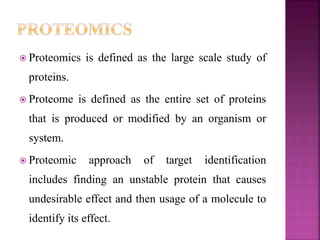
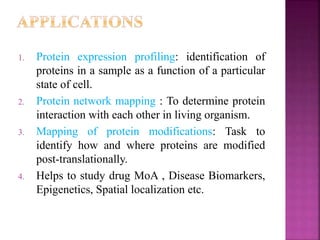
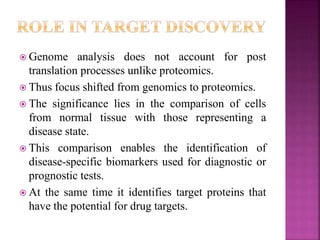
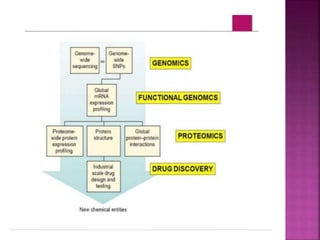
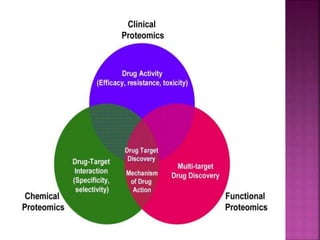
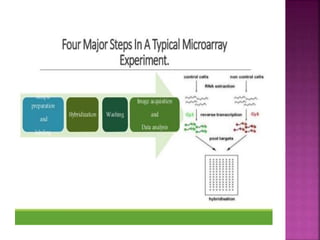
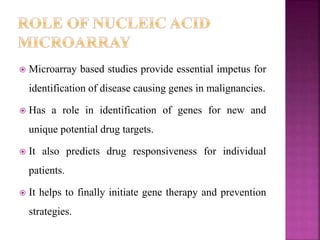
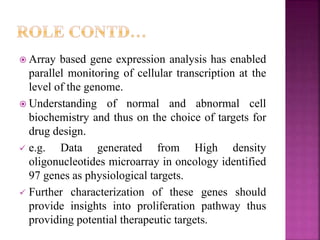
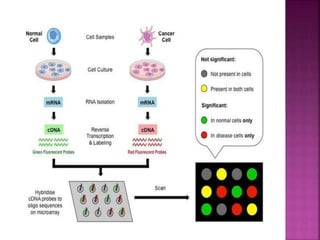
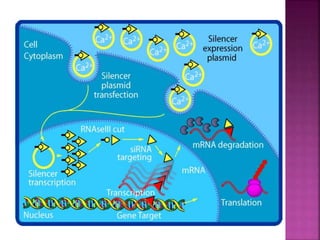
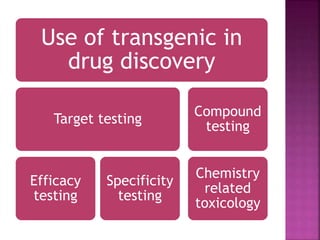
The document discusses drug discovery processes, focusing on target identification and validation through various methods including high-throughput screening (HTS), genomics, proteomics, and bioinformatics. It highlights the significance of understanding molecular targets related to diseases, utilizing transgenic animals for efficacy and specificity testing, and employing techniques such as microarrays and antisense technology. Overall, it emphasizes the integration of multiple scientific approaches to improve drug discovery and development outcomes.