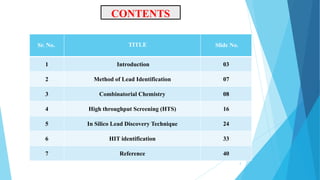
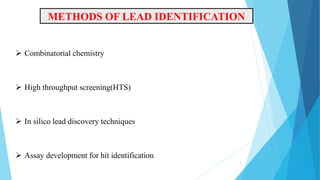
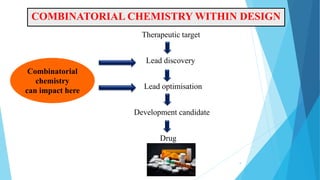
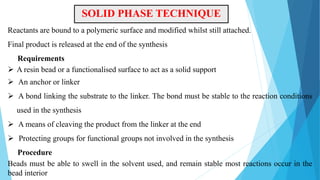
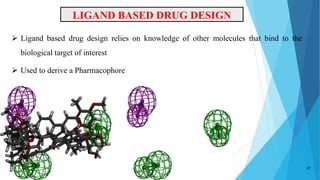
This document provides an overview of lead identification in drug discovery. It discusses various methods for identifying lead compounds, including combinatorial chemistry, high-throughput screening, and in silico lead discovery techniques. Combinatorial chemistry allows for the rapid production and screening of large compound libraries. High-throughput screening assays test large numbers of compounds against biological targets using automated technologies. In silico methods like molecular docking use computer simulations to predict how compounds may bind and interact with targets. The goal is to find initial "hit" compounds that can then be optimized into drug candidates.


























![MOLECULAR DOCKING
In the field of molecular modeling, docking is a method which predicts the preferred
orientation of one molecule to a second when bound to each other to form a stable complex.[1]
Knowledge of the preferred orientation in turn may be used to predict the strength of
association or binding affinity between two molecules using, for example, scoring functions.
32](https://image.slidesharecdn.com/suhas2ndsemseminarfinalwork-200212083943/85/LEAD-IDENTIFICATION-BY-SUHAS-PATIL-S-K-32-320.jpg)








