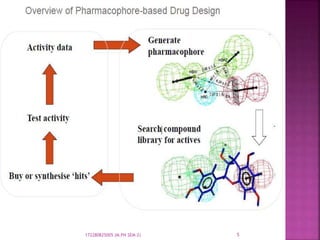
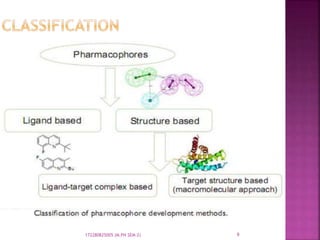
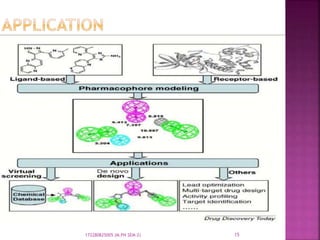
The document discusses pharmacophores, which are abstract descriptions of molecular features necessary for molecular recognition between a ligand and biological macromolecule. A pharmacophore consists of 3D structural features like hydrophobic groups and hydrogen bond donors/acceptors. Pharmacophore mapping is used to define pharmacophoric features and align molecules to identify common binding elements responsible for biological activity. Pharmacophore models can be used in virtual screening to filter large databases and identify new compounds that may bind similarly to known active molecules. The document provides details on different approaches for pharmacophore generation and searching compound libraries.