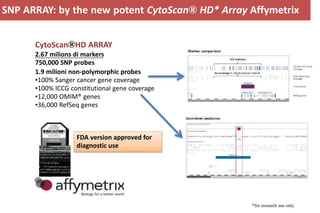
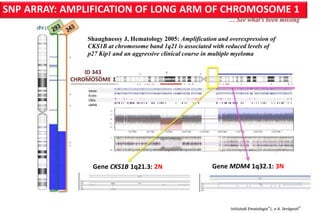
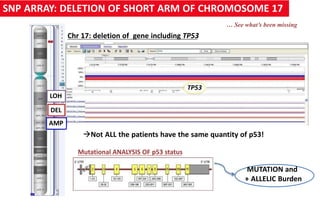
The document discusses the application of Affymetrix arrays in studying hematological malignancies, focusing on the detection of genetic alterations associated with acute promyelocytic leukemia (APL) through high-resolution single nucleotide polymorphism (SNP) arrays. It highlights the significance of identifying actionable cancer biomarkers for personalized therapy and presents data on genomic profiling, copy number alterations, and gene expression analysis in different patient cohorts. Furthermore, it outlines the implications of genetic findings on prognosis and treatment strategies for acute leukemia patients.