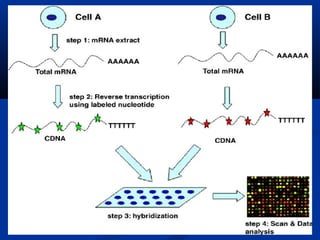
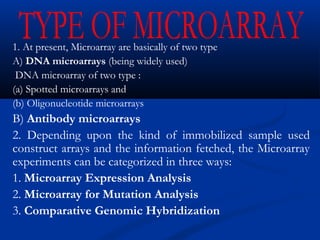
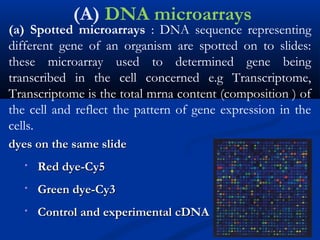
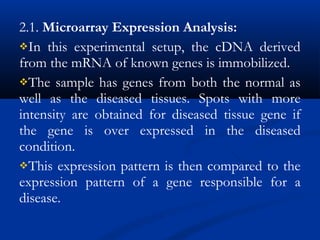
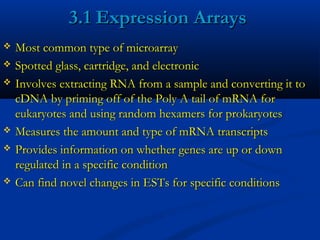
This document discusses the significance and advancements of DNA microarray technology in molecular biology, enabling the analysis of multiple genes simultaneously to understand gene expression, genetic disorders, and disease mechanisms. It covers the methodology of microarray experiments, different types of microarrays, and their applications in gene discovery, disease diagnosis, and drug development. The technology allows for rapid and sensitive analysis of gene expression profiles, offering insights into various biological processes.





![3. Other Types of microarrays include:
1) Expression array
2) Protien microarray
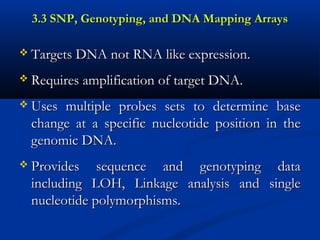
3) SNP, Genotyping, and DNA Mapping ArraysSNP, Genotyping, and DNA Mapping Arrays
4) Resequencing Arrays [Affy]4) Resequencing Arrays [Affy]
5)5) Tissue microarrays
6) Cellular microarrays (also called transfection
microarrays)
7) Chemical compound microarrays
8) Carbohydrate arrays (glycoarrays)
9) Phenotype microarrays
10) Reverse Phase Protein Microarrays](https://image.slidesharecdn.com/seminar-sumantsingh621-180103164404/85/A-comprehensive-study-of-microarray-11-320.jpg)



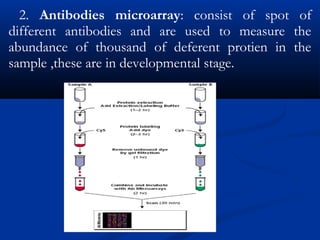
![3.2 Protein Microarrays3.2 Protein Microarrays
True protein microarrays are evolving very slowTrue protein microarrays are evolving very slow
and only a few exist.and only a few exist.
Technology is not straight forward due toTechnology is not straight forward due to
inherent characteristic of proteins [e.g. availableinherent characteristic of proteins [e.g. available
ligands, folding, drying…]ligands, folding, drying…]
Most are designed to detect antibodies orMost are designed to detect antibodies or
enzymes in a biological systemenzymes in a biological system
Protein is on the microarrayProtein is on the microarray
Some detect protein-protein interaction bySome detect protein-protein interaction by
surface plasmon resonance other use asurface plasmon resonance other use a
fluorescence based approachfluorescence based approach](https://image.slidesharecdn.com/seminar-sumantsingh621-180103164404/85/A-comprehensive-study-of-microarray-20-320.jpg)
![3.4 Resequencing Arrays [Affy]3.4 Resequencing Arrays [Affy]
Enable the analysis of up to 300,000+ bases ofEnable the analysis of up to 300,000+ bases of
double-stranded sequence (600,000 bases total)double-stranded sequence (600,000 bases total)
on a single Affy array.on a single Affy array.
Used for large-scale resequencing of organismsUsed for large-scale resequencing of organisms
genome and organelles.genome and organelles.
Faster and cheaper than sequencing but veryFaster and cheaper than sequencing but very
limited to few organisms and/or organelles.limited to few organisms and/or organelles.
Large potential.Large potential.](https://image.slidesharecdn.com/seminar-sumantsingh621-180103164404/85/A-comprehensive-study-of-microarray-22-320.jpg)