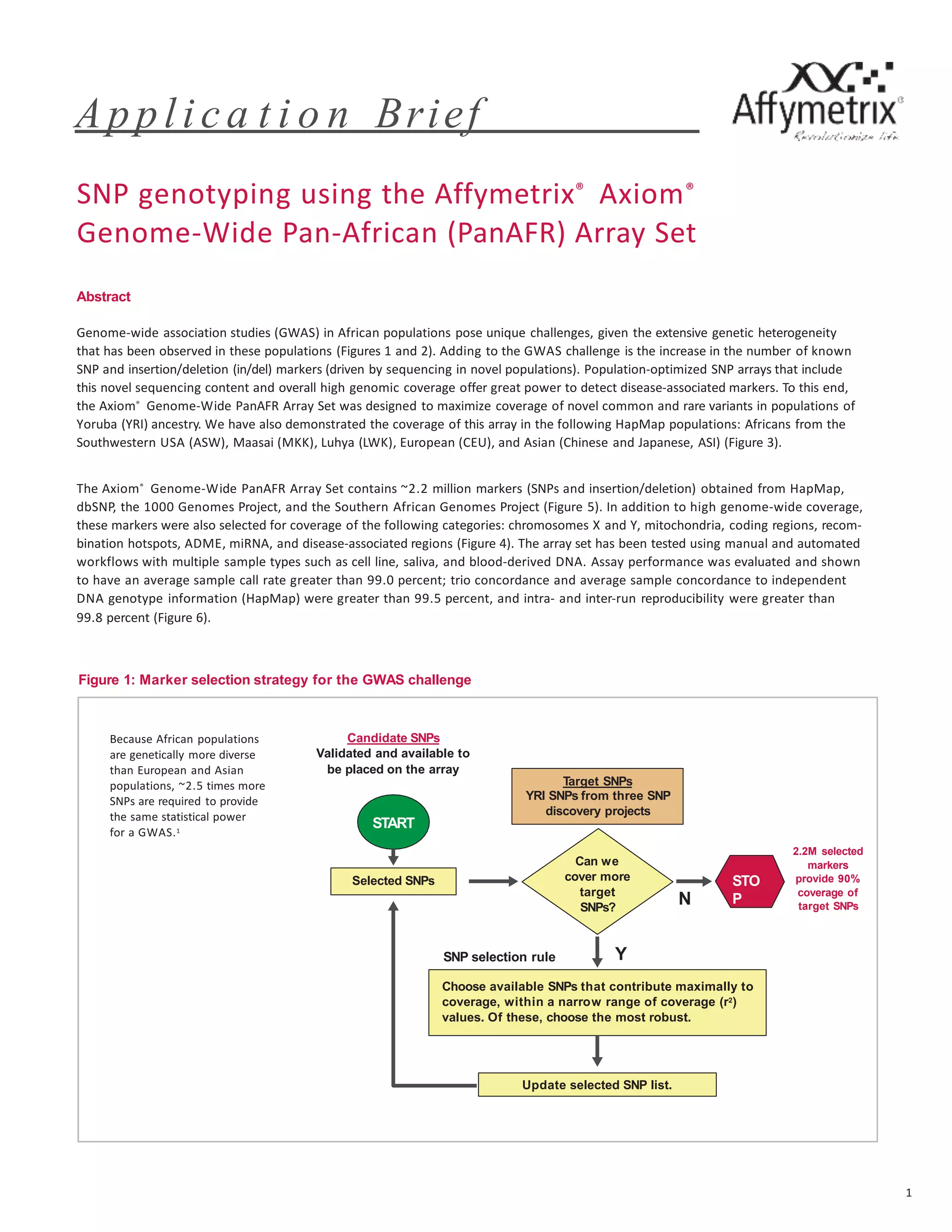
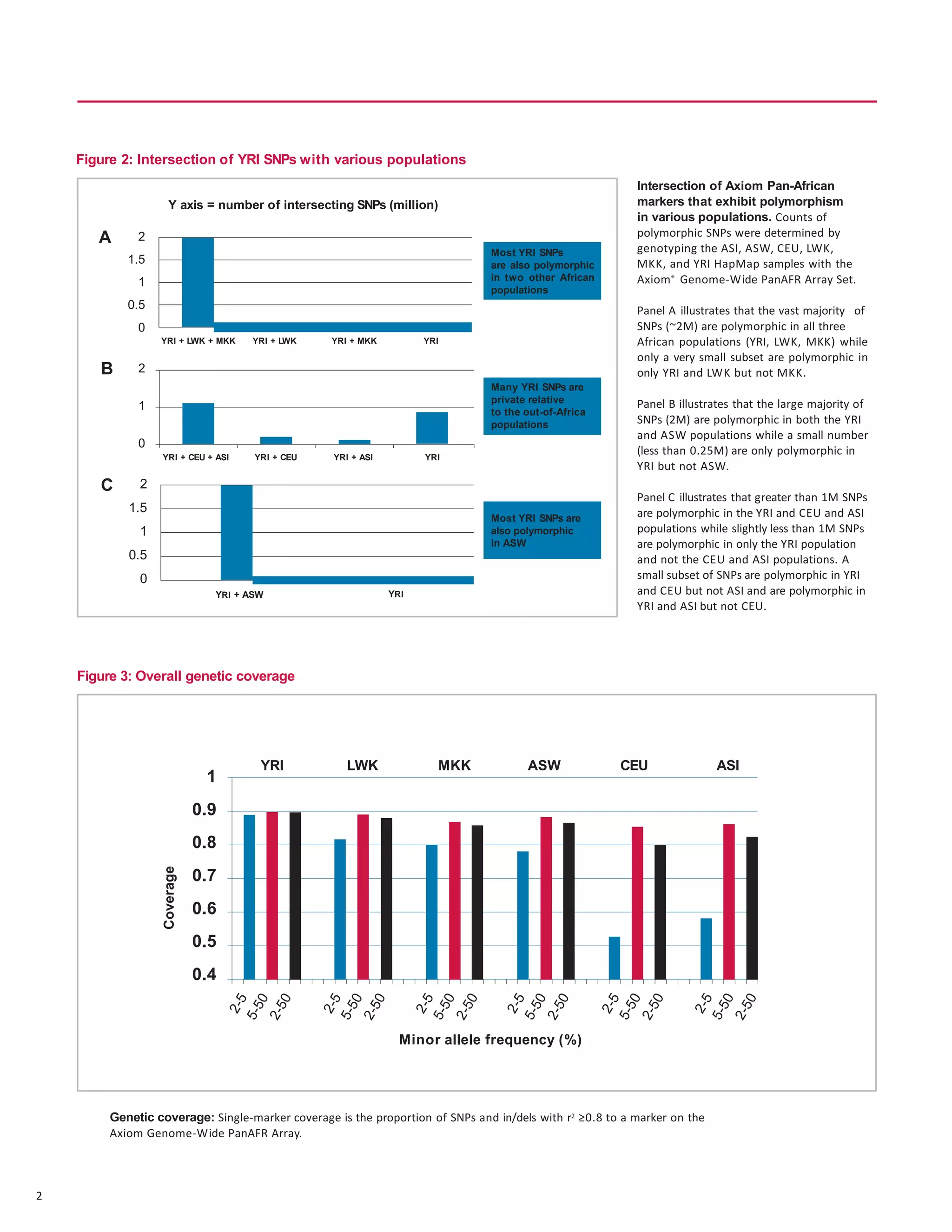
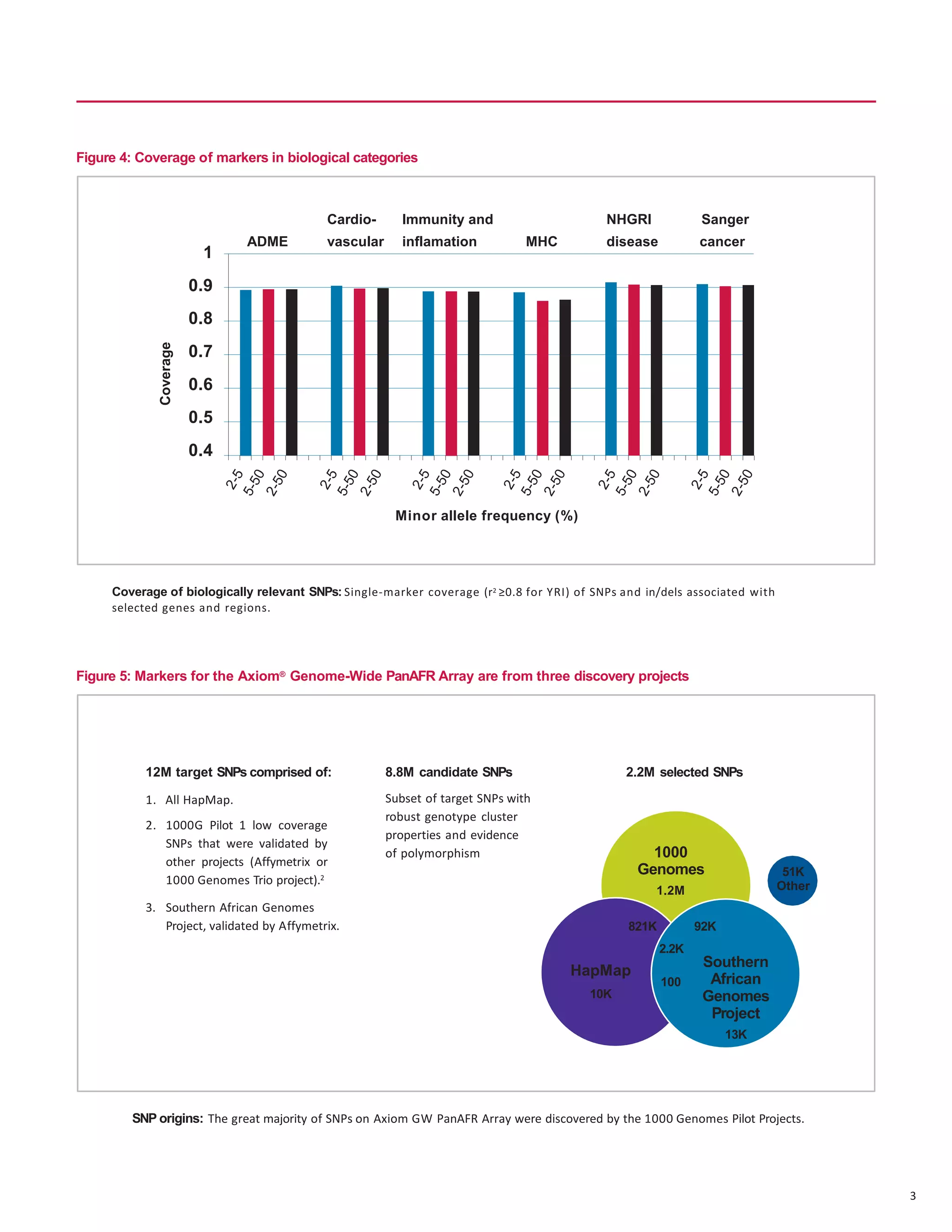
The document describes the Axiom Genome-Wide PanAFR Array Set, which was designed to maximize coverage of common and rare genetic variants in populations of Yoruba ancestry. The array contains over 2.2 million markers selected from several genome projects to provide high coverage of the genome as well as specific regions and gene categories. Testing showed the array performs well with an average call rate over 99% and high concordance and reproducibility of calls.