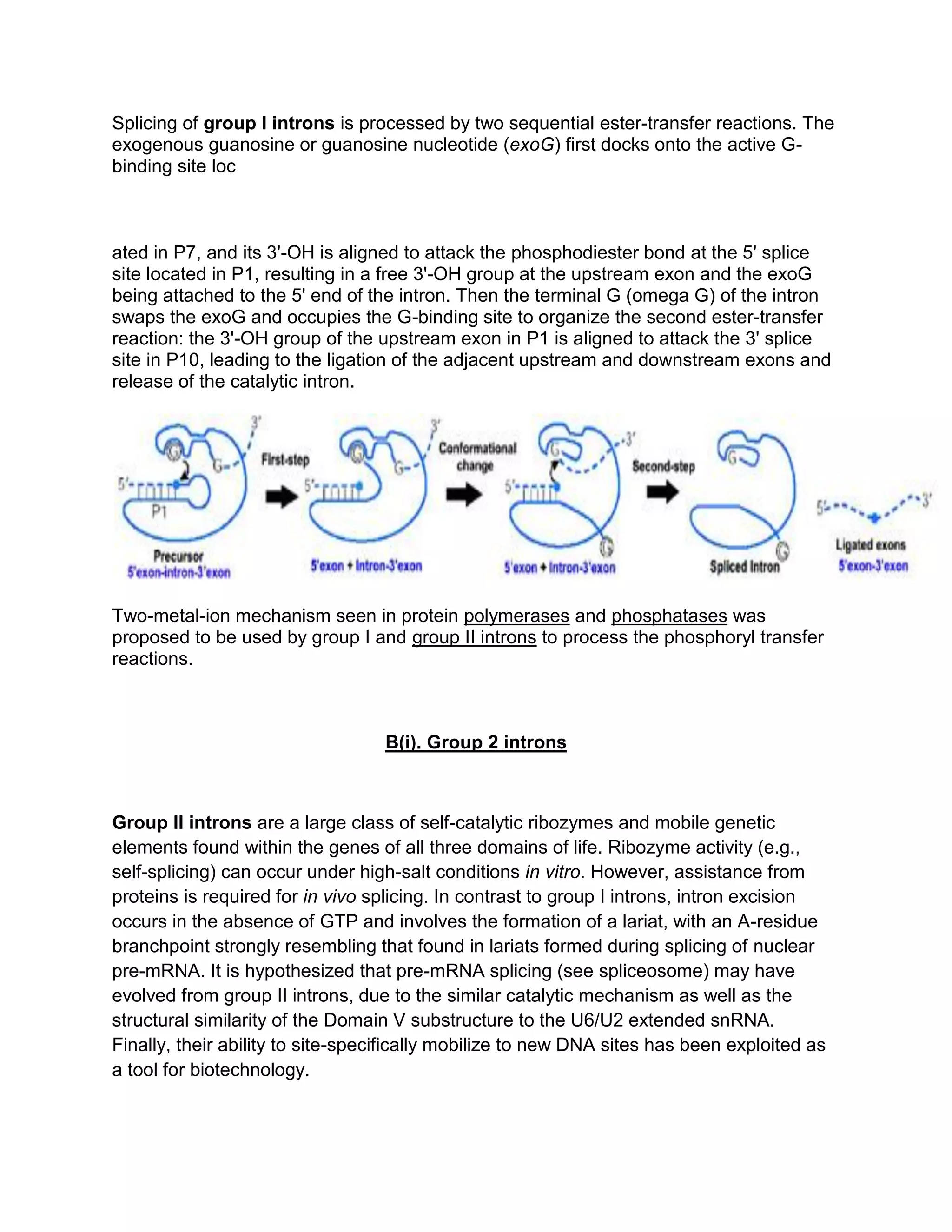
Group I introns are self-splicing ribozymes that catalyze their own excision from mRNA, tRNA, and rRNA precursors. They consist of a conserved core of nine paired regions (P1-P9) that fold into two domains. Splicing occurs via two sequential transesterification reactions involving the docking of an exogenous guanosine cofactor.
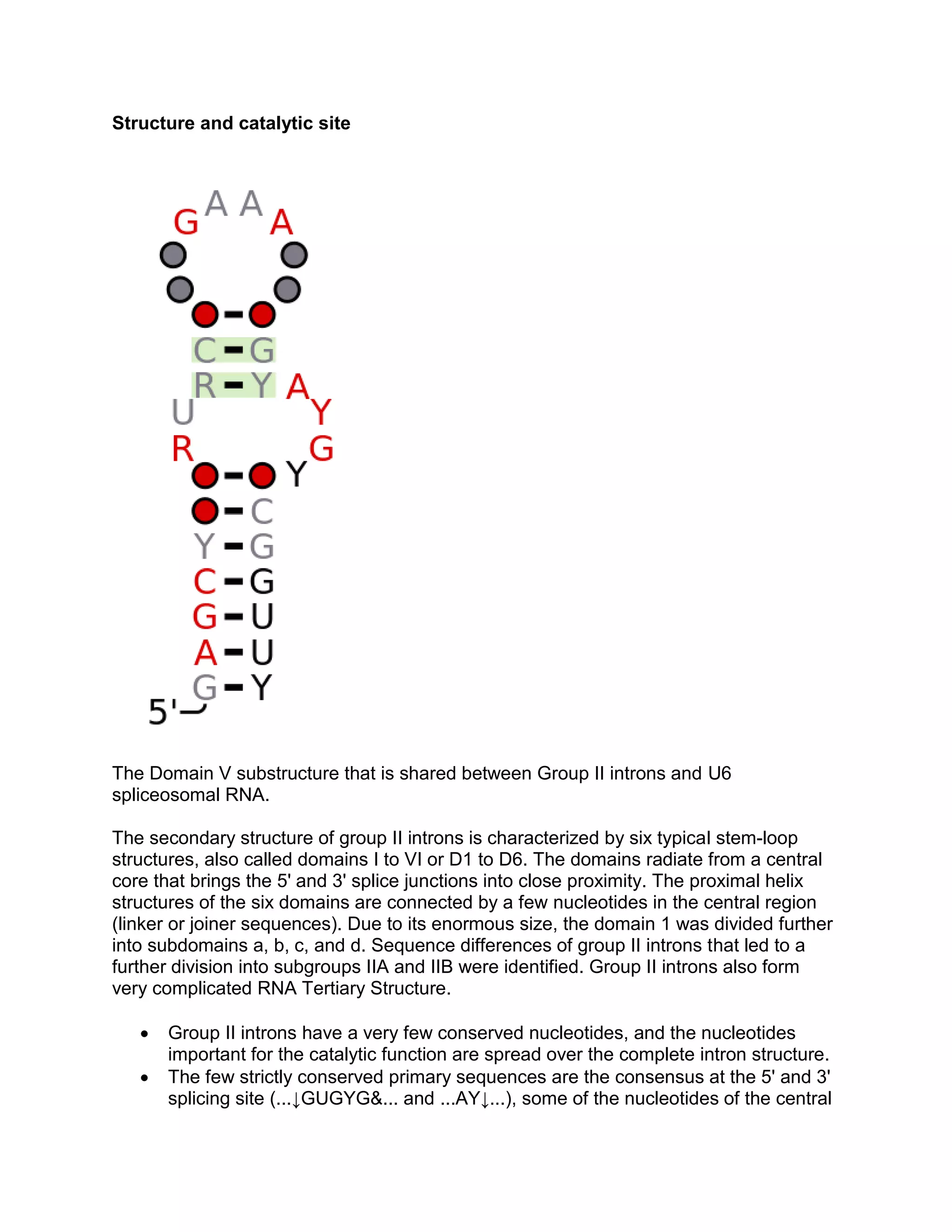
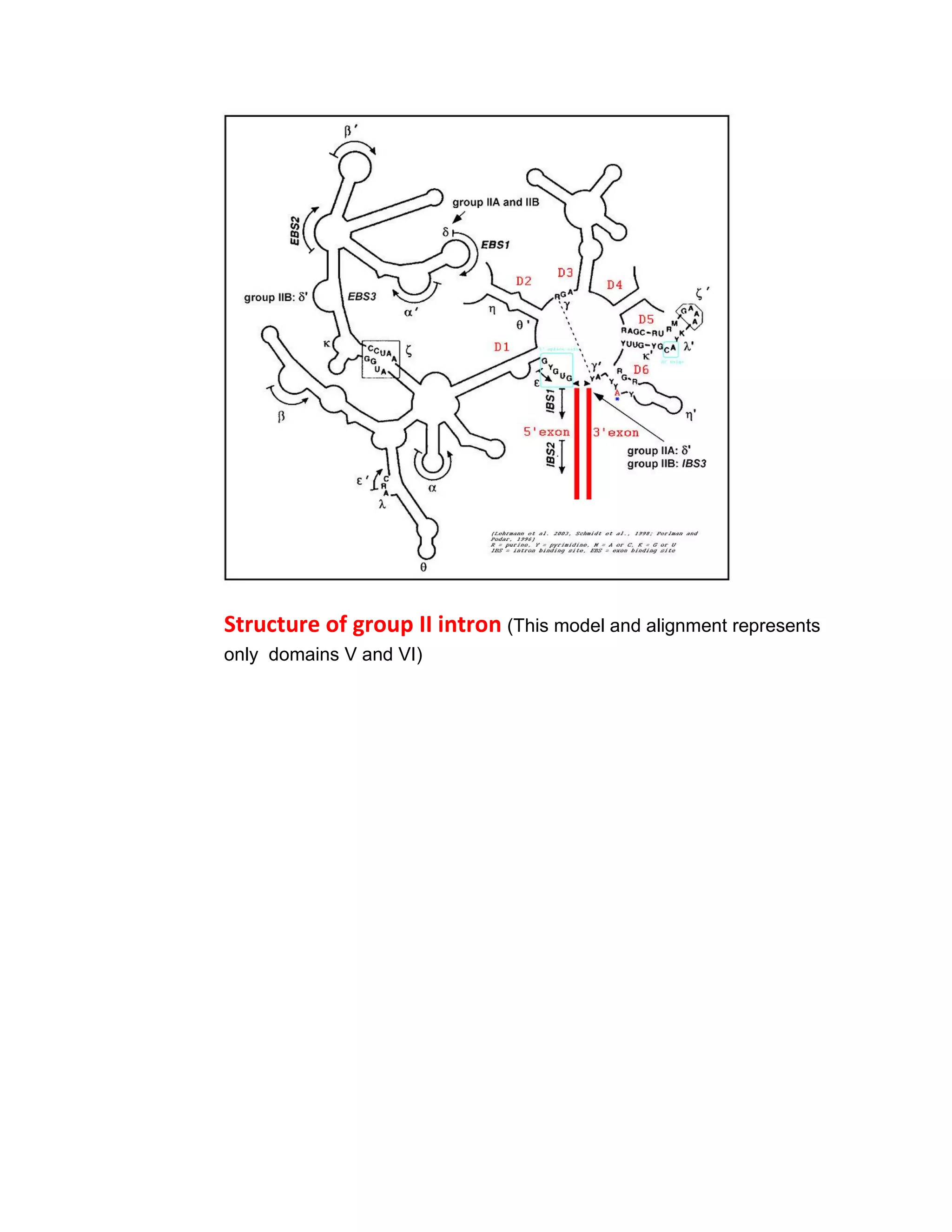
Group II introns are self-splicing ribozymes and mobile genetic elements found in all domains of life. They consist of six stem-loop domains (I-VI) radiating from a central core. Splicing occurs via two steps like pre-mRNA splicing, forming a lariat intermediate with a branchpoint