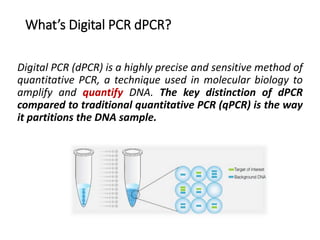
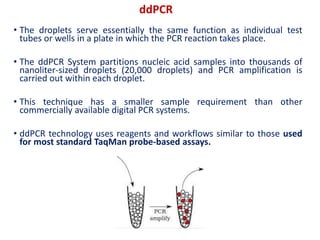
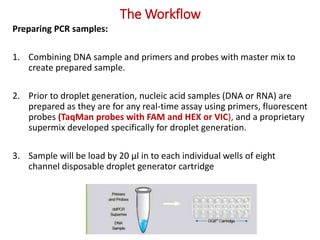
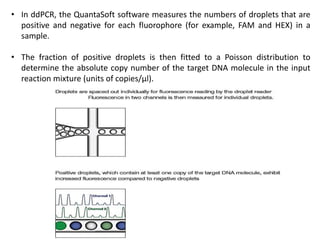
Digital PCR (dPCR) is a highly sensitive technique that partitions a DNA sample into thousands of individual reactions to quantify DNA molecules. Droplet digital PCR (ddPCR) further partitions samples into nanoliter-sized droplets in oil emulsion. ddPCR provides absolute quantification without standard curves by counting positive and negative droplets. It has advantages over qPCR including detection of rare sequences, single cell analysis, and use in validating next-generation sequencing.