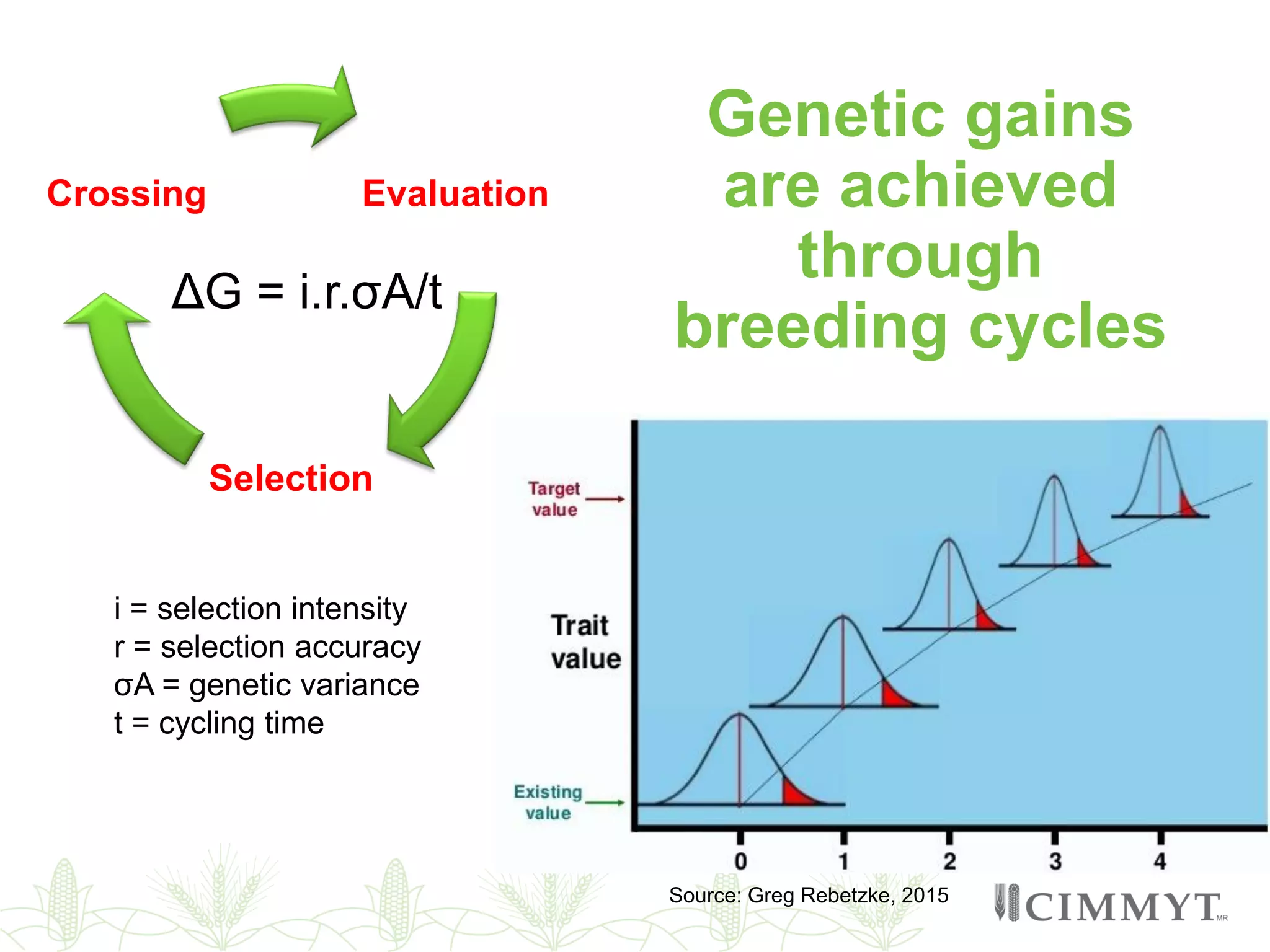
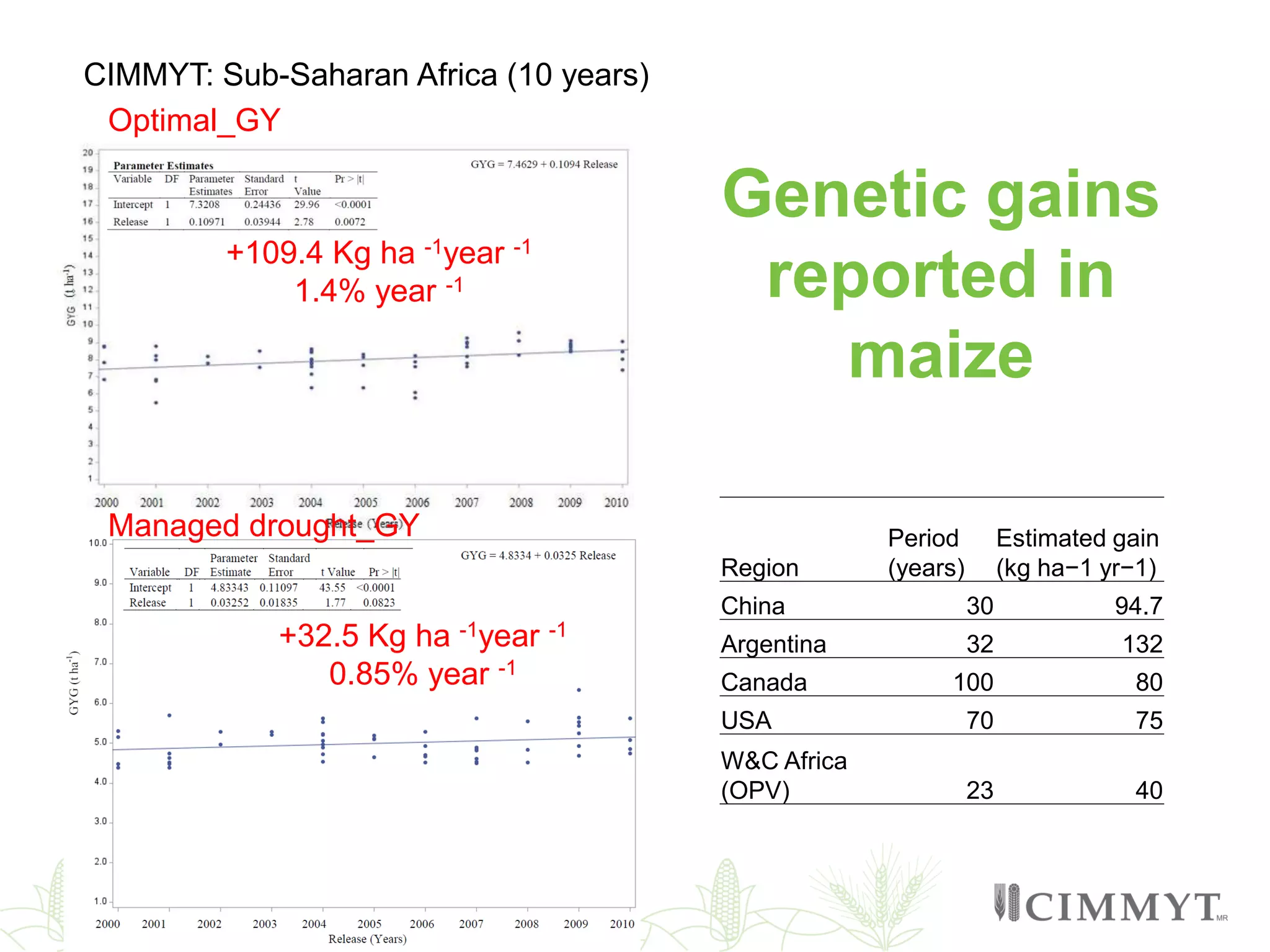
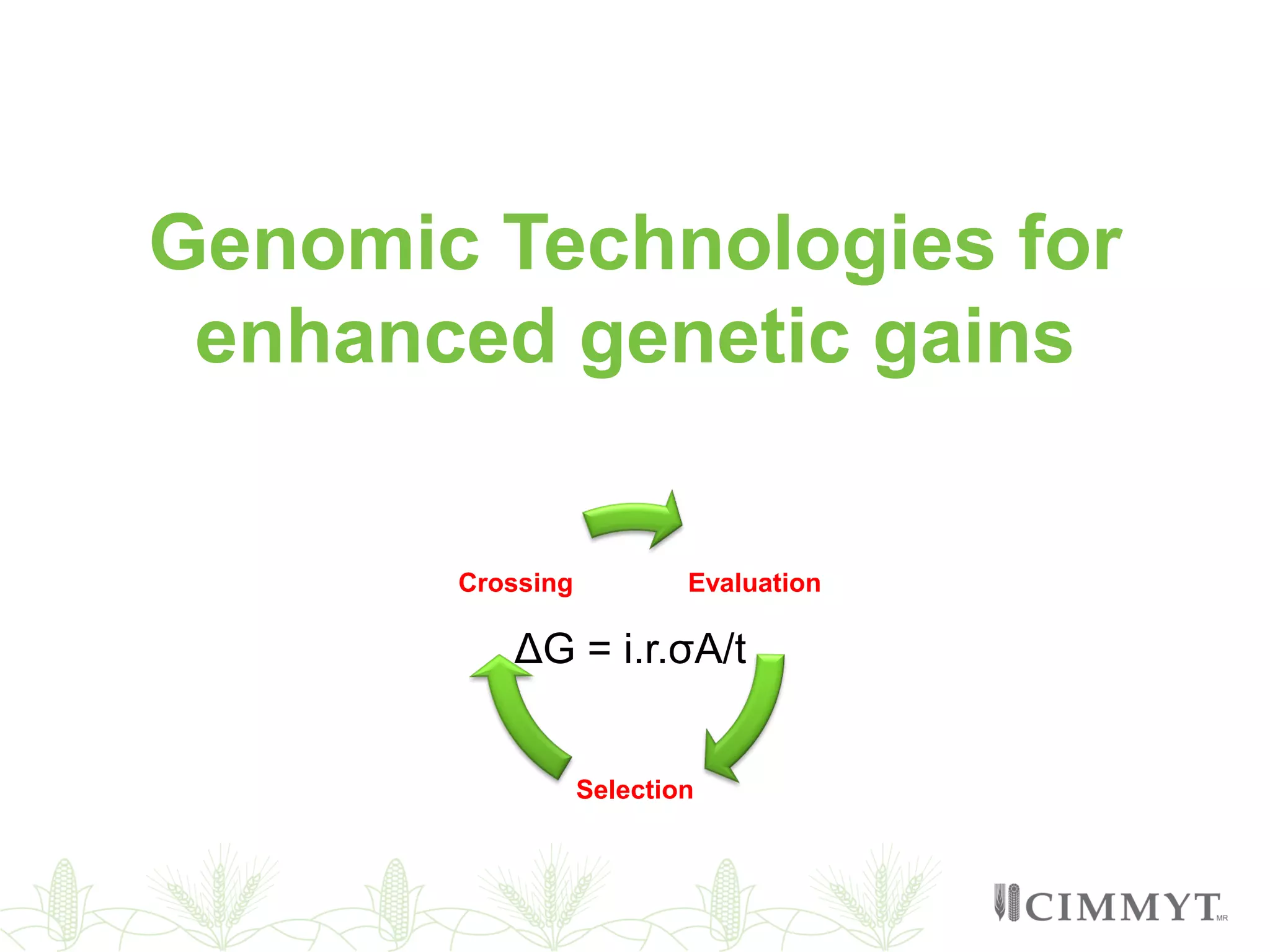
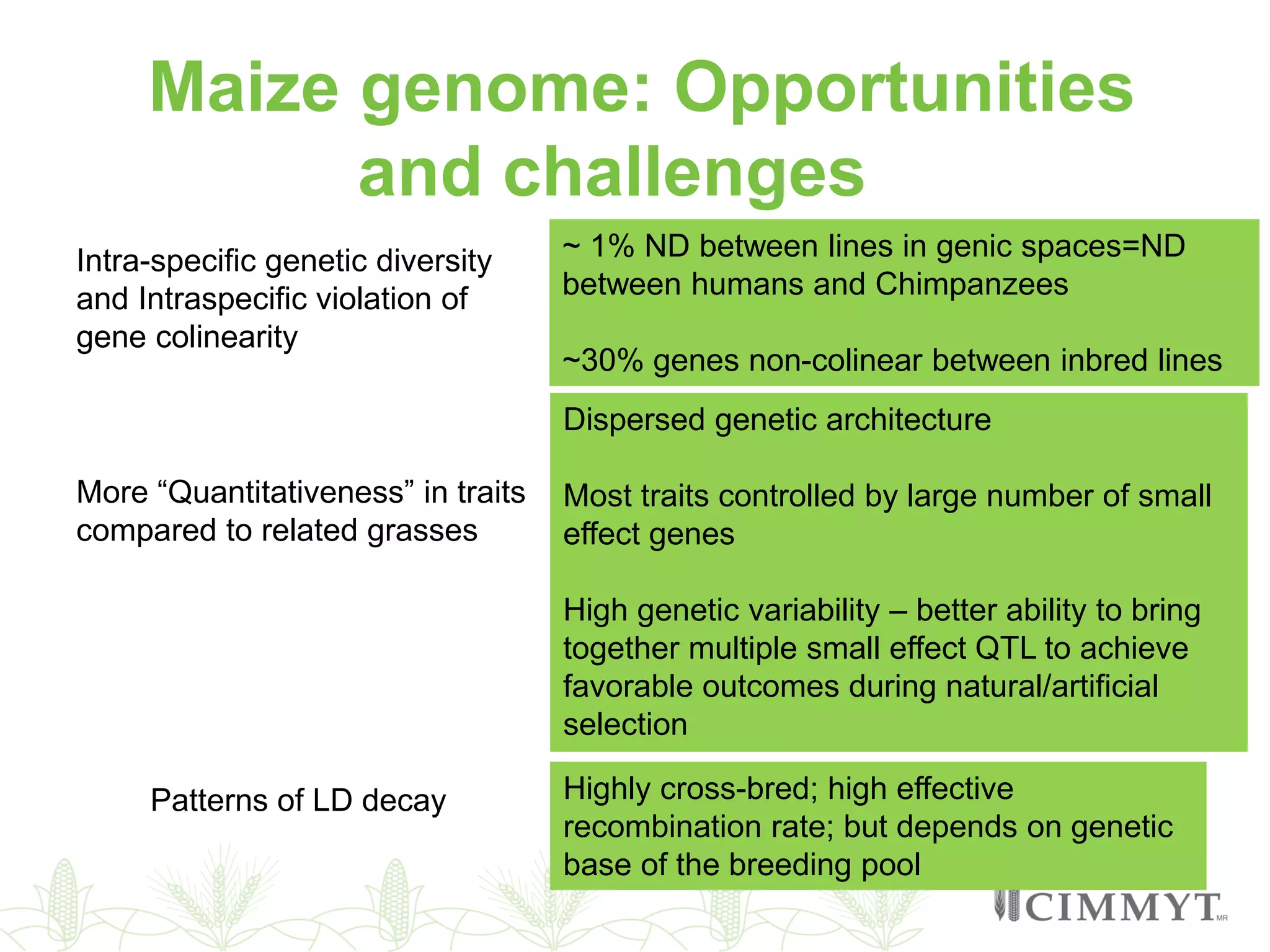
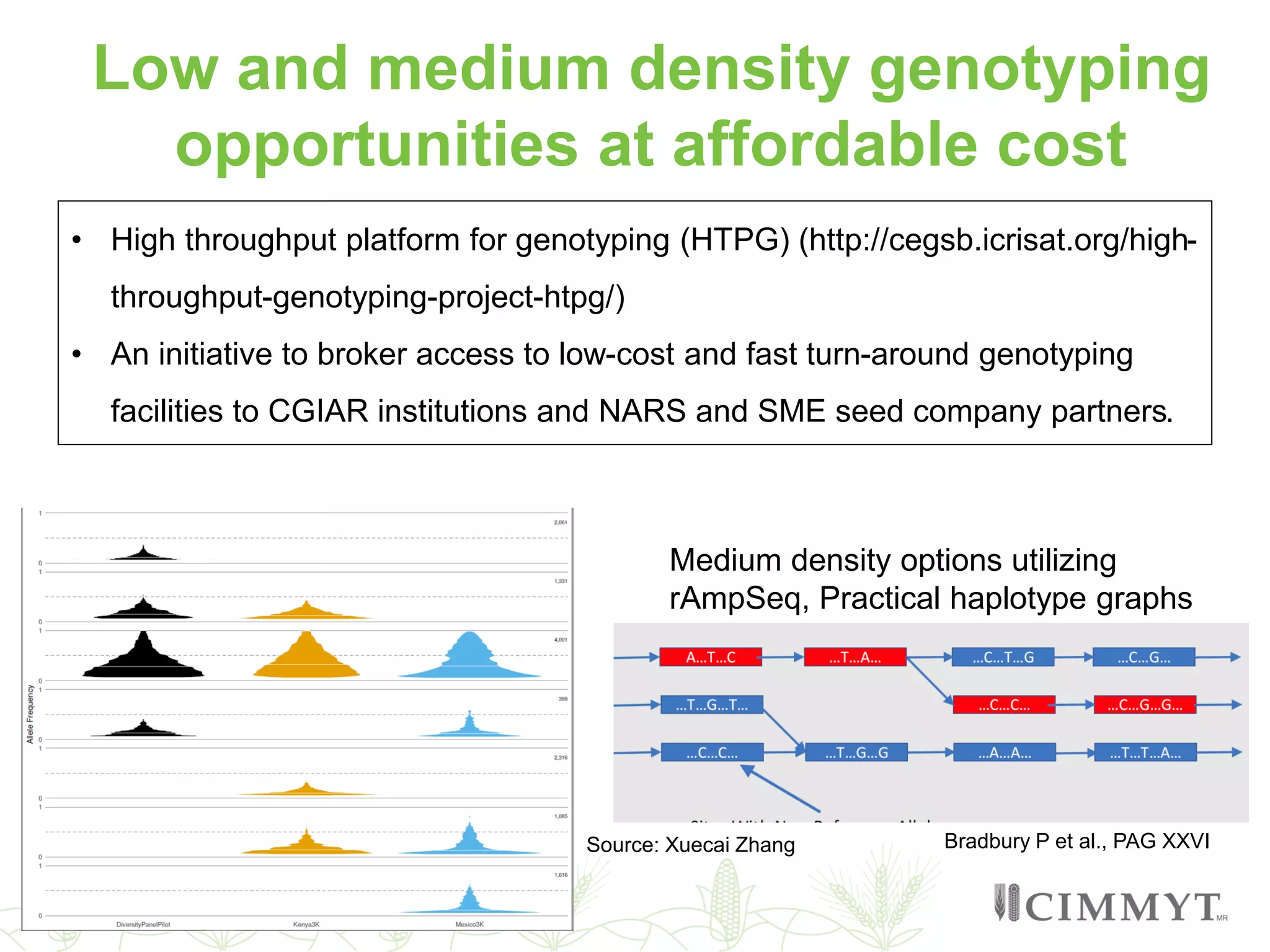
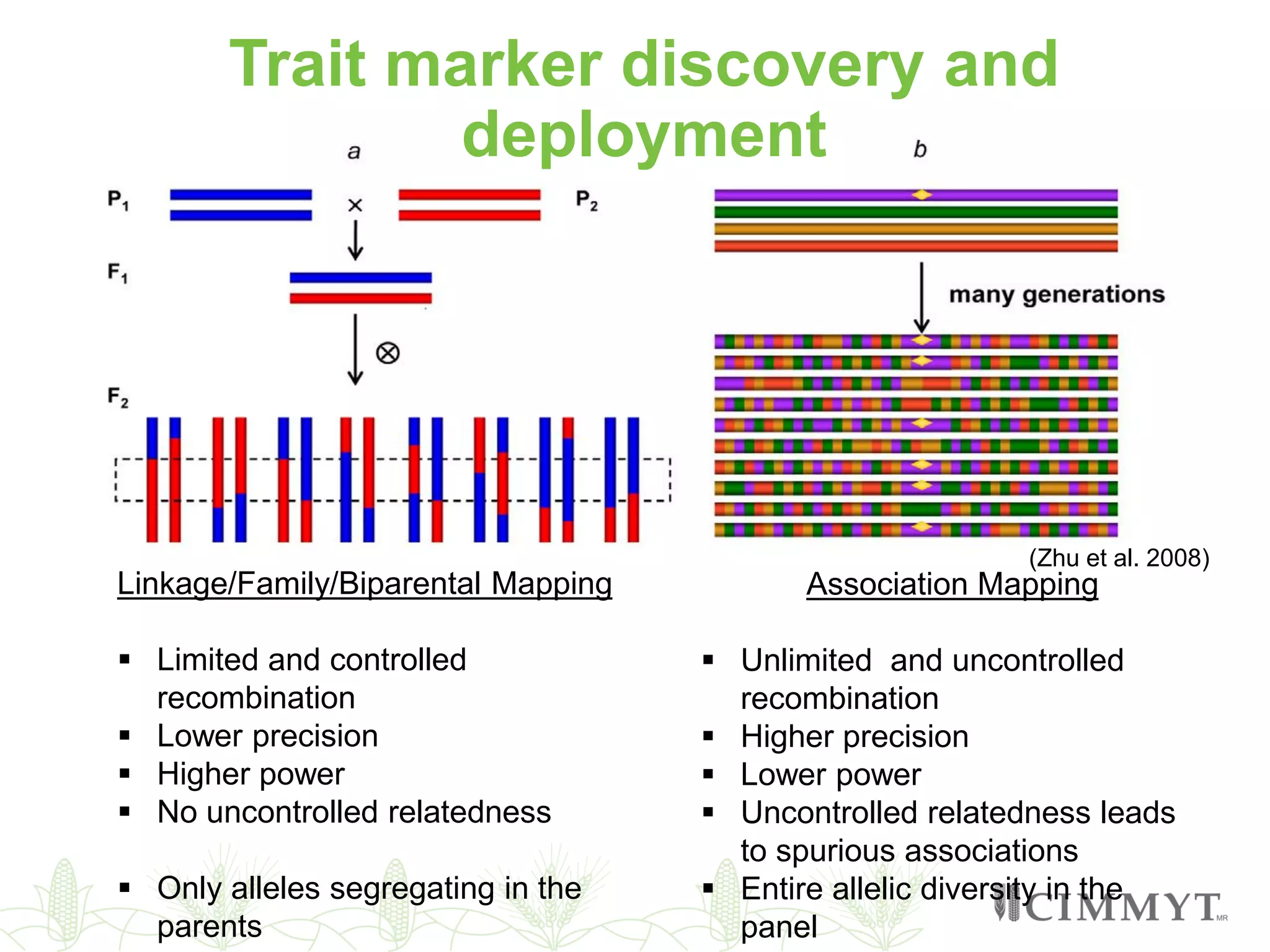
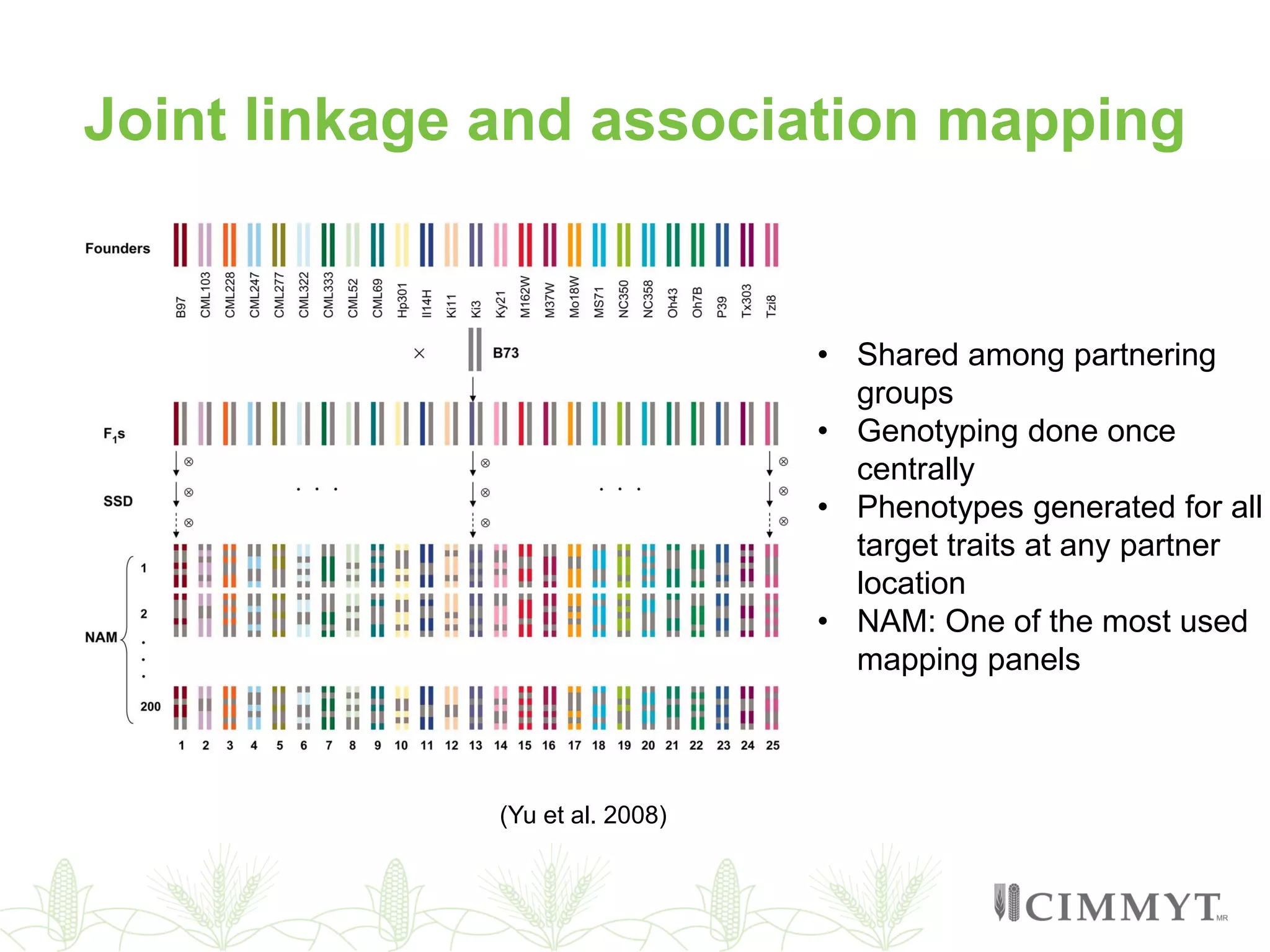
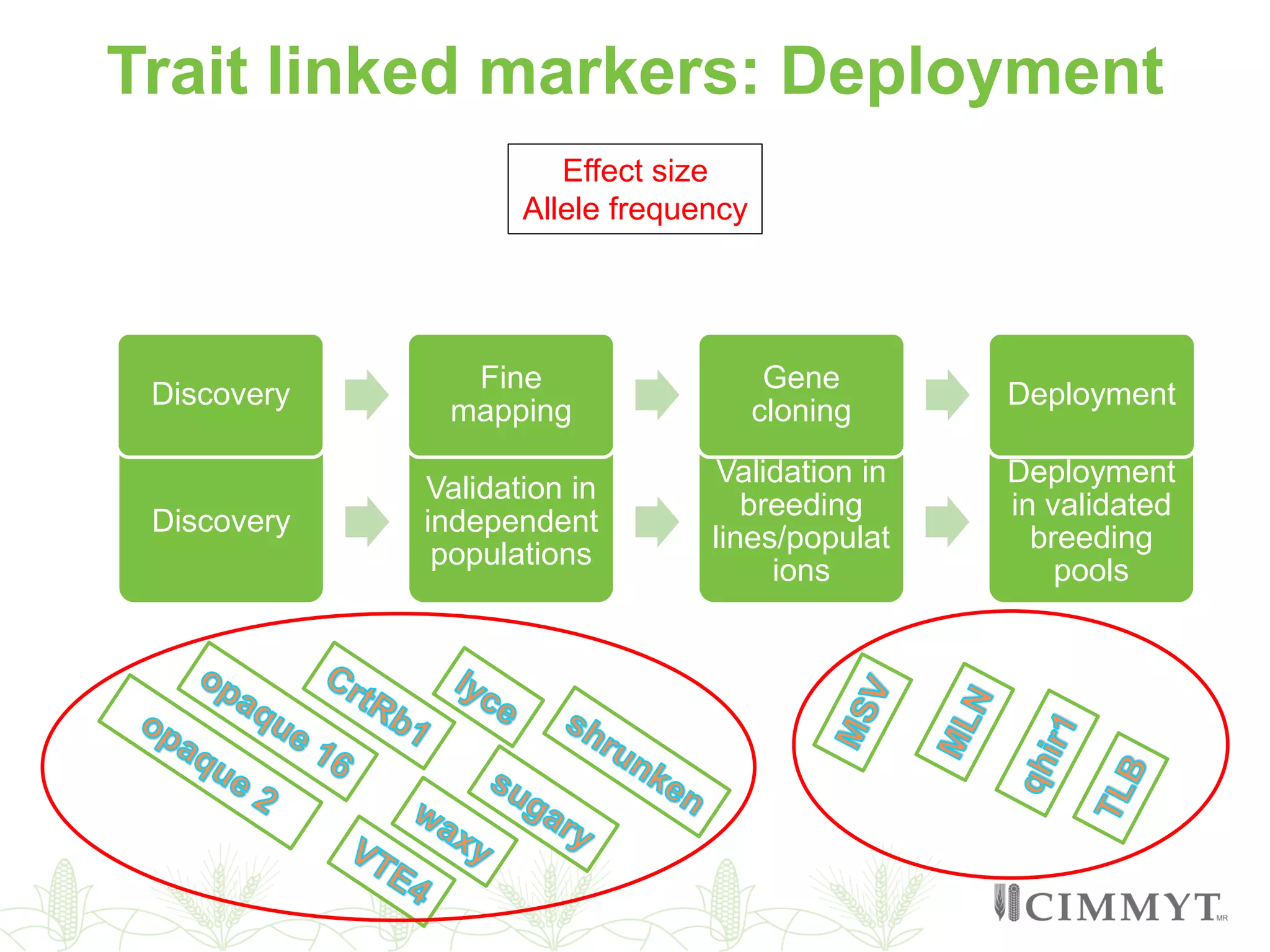
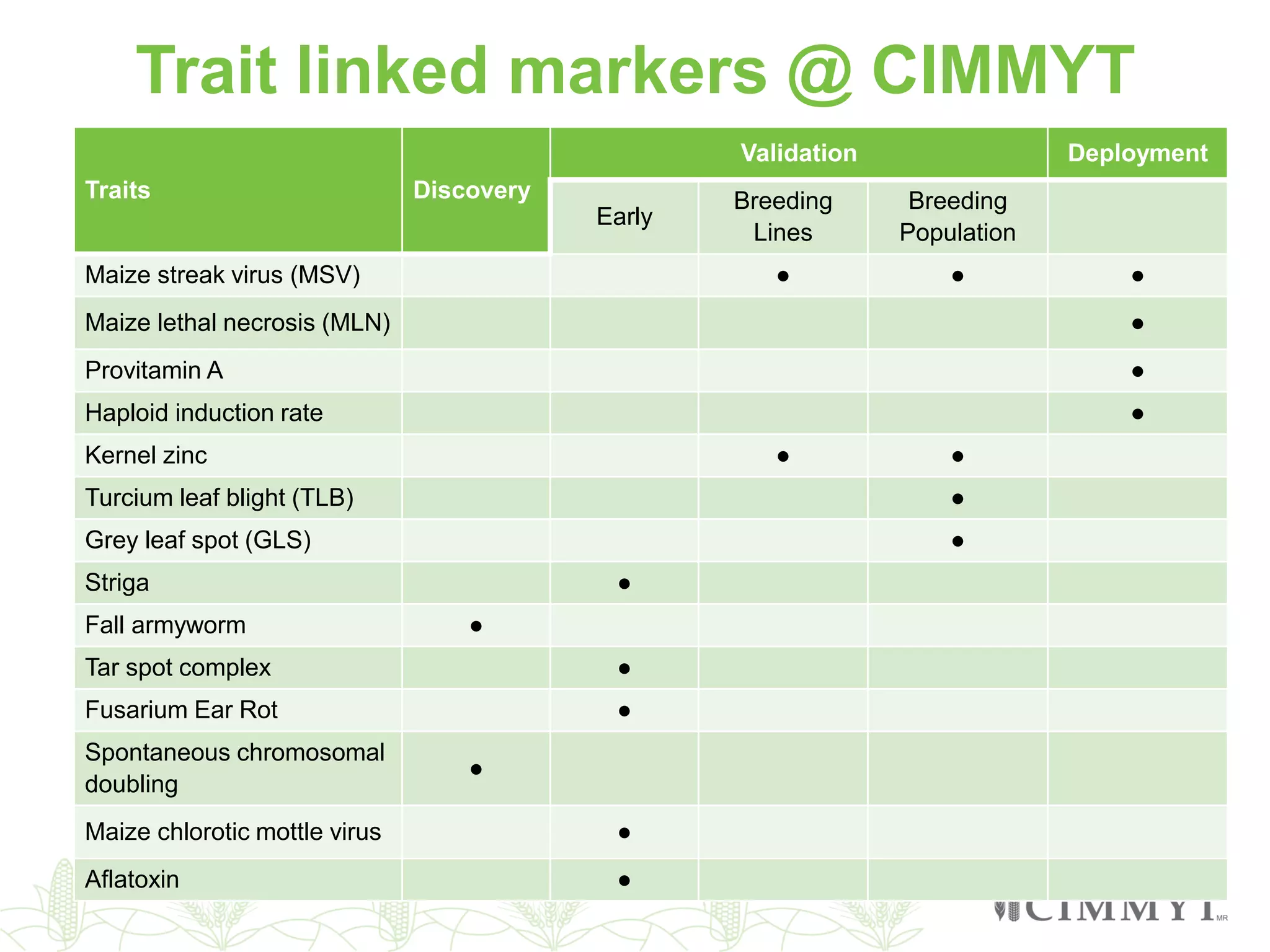
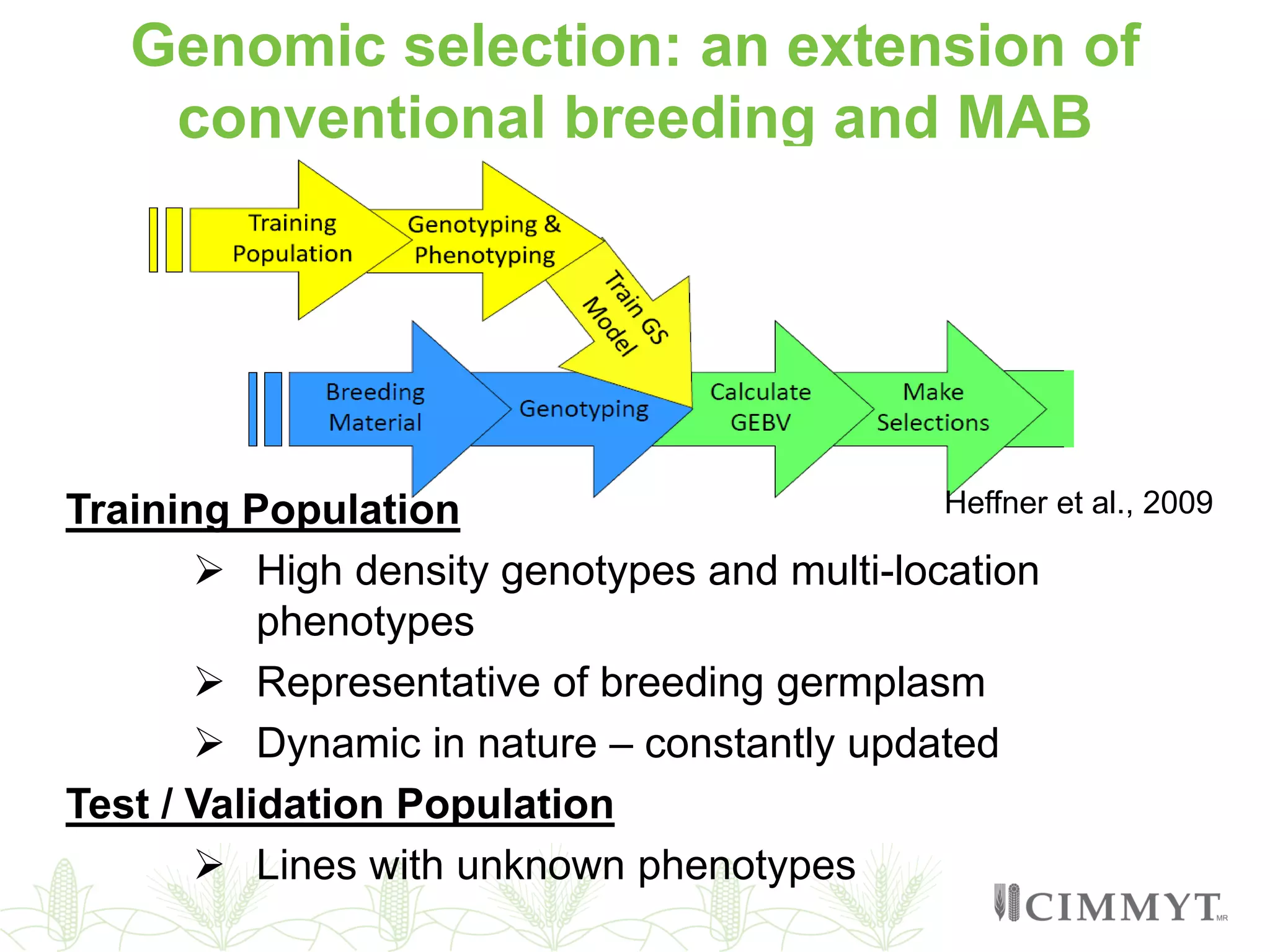
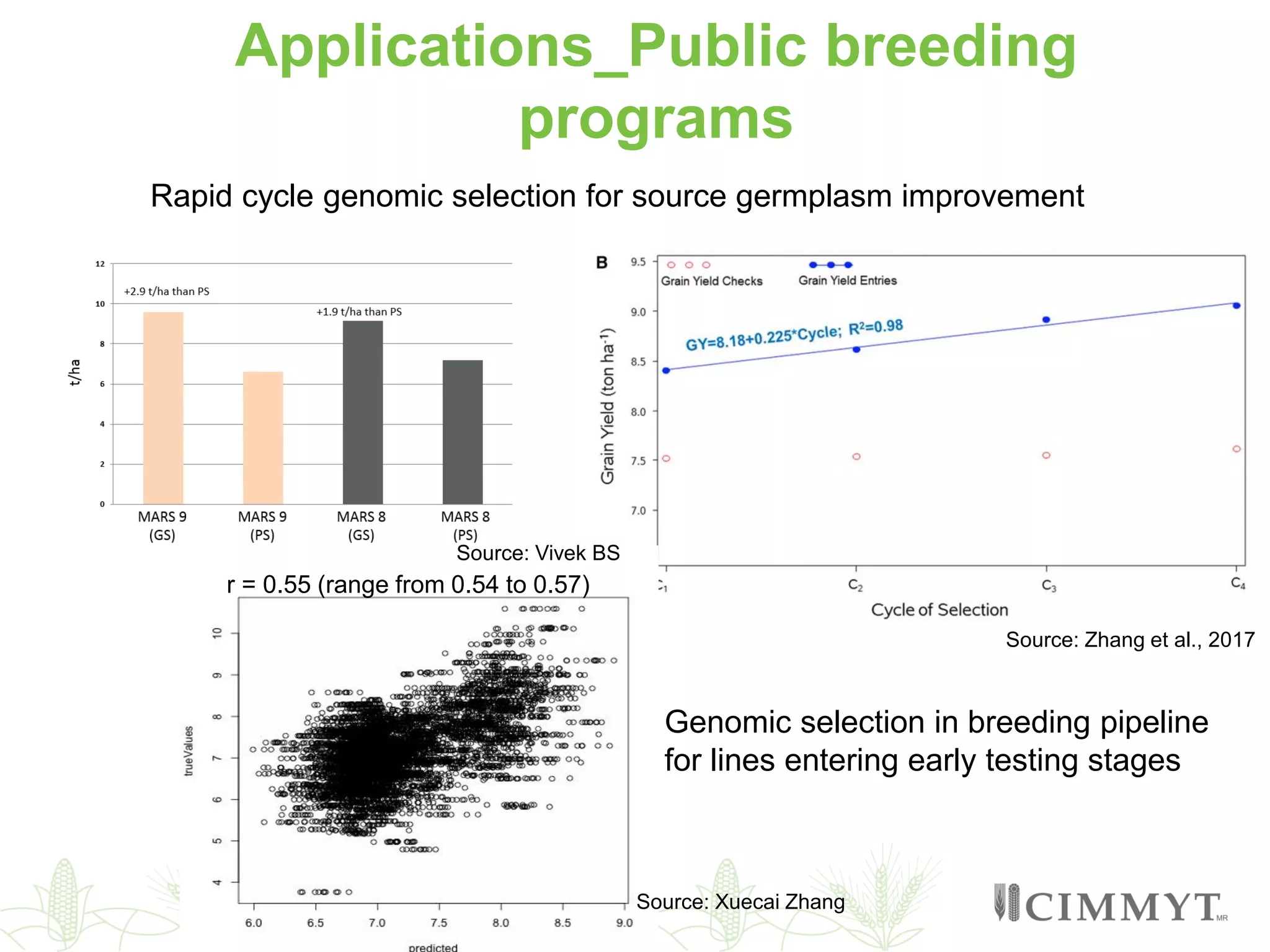
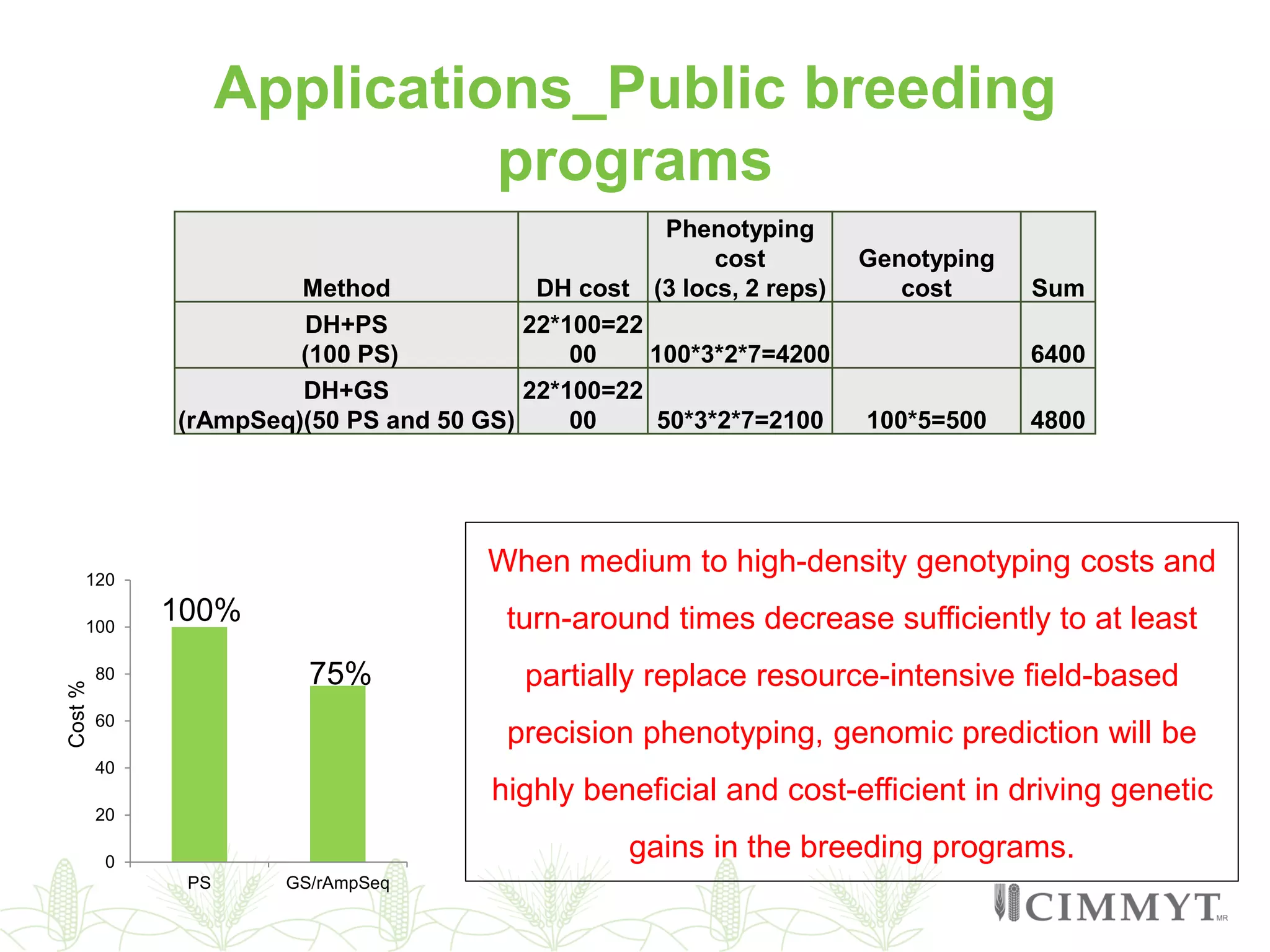
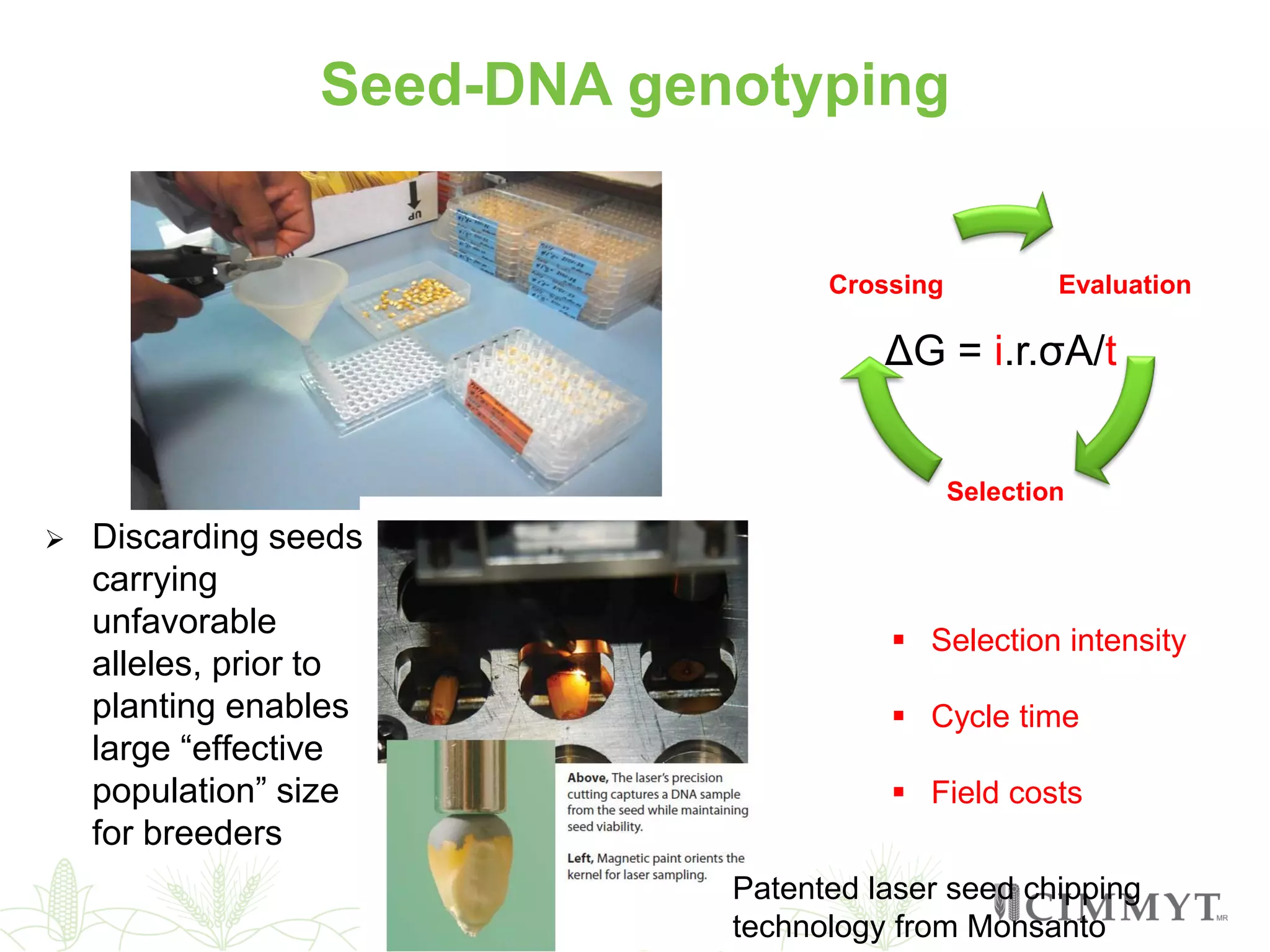
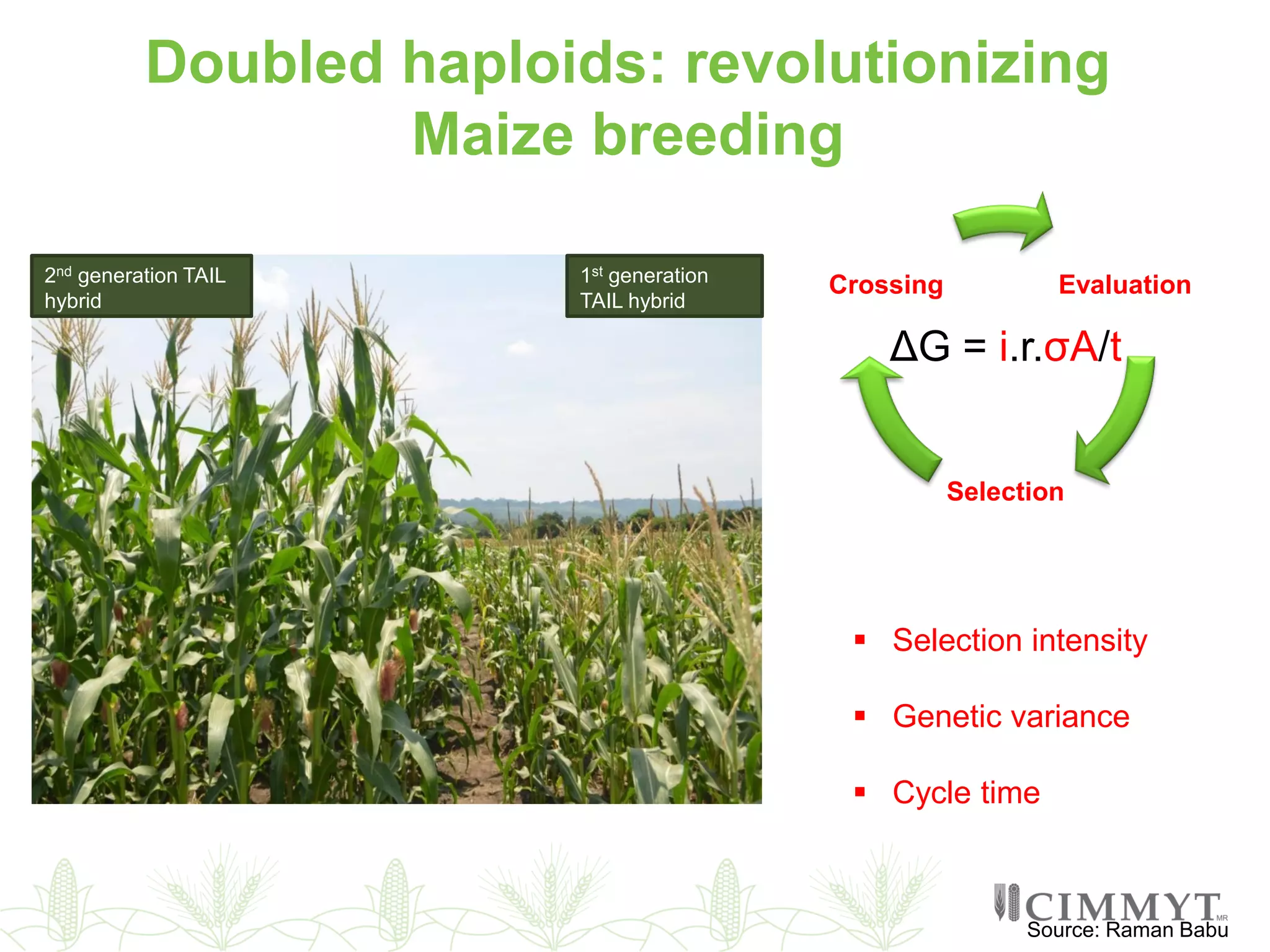
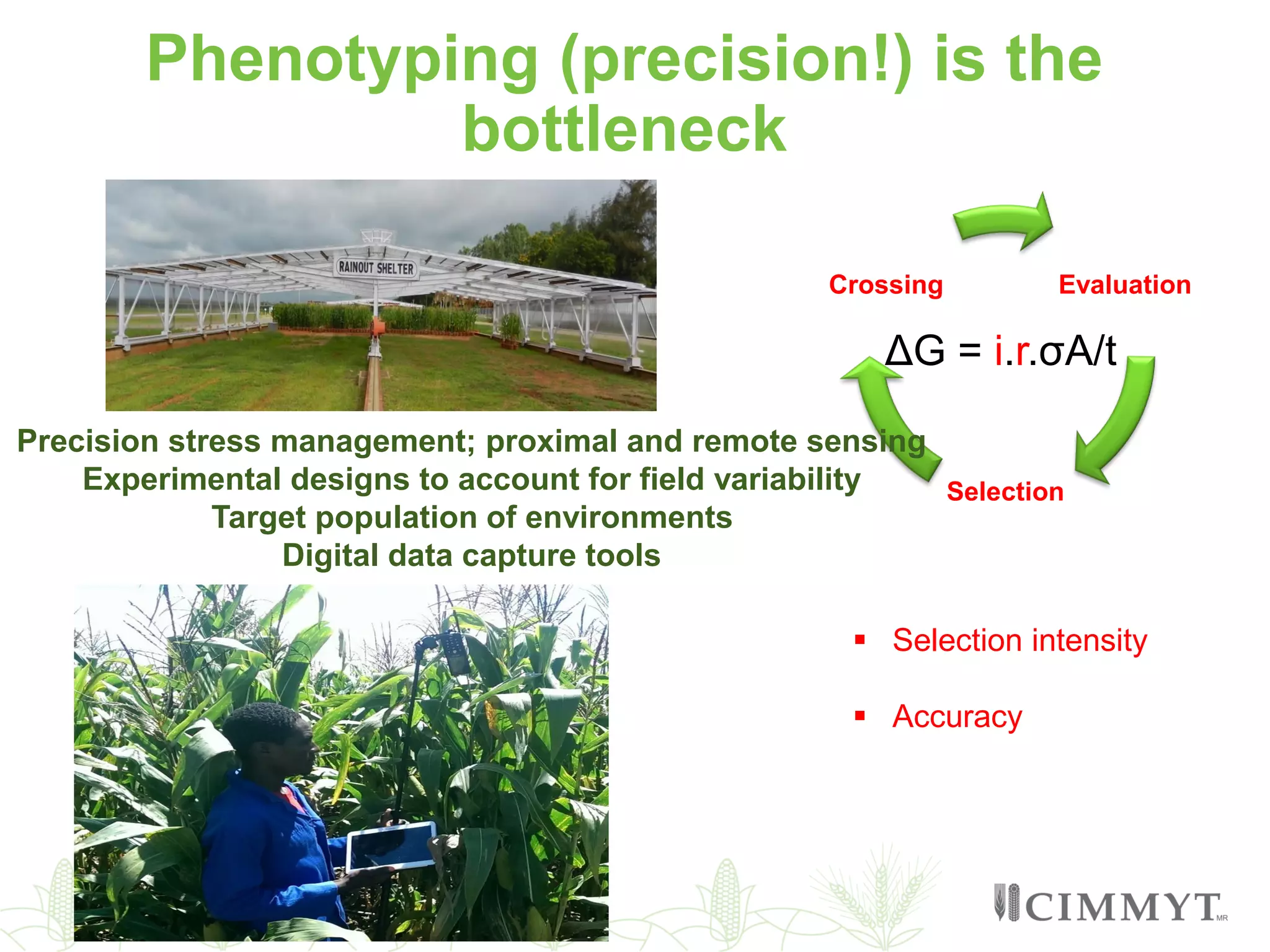
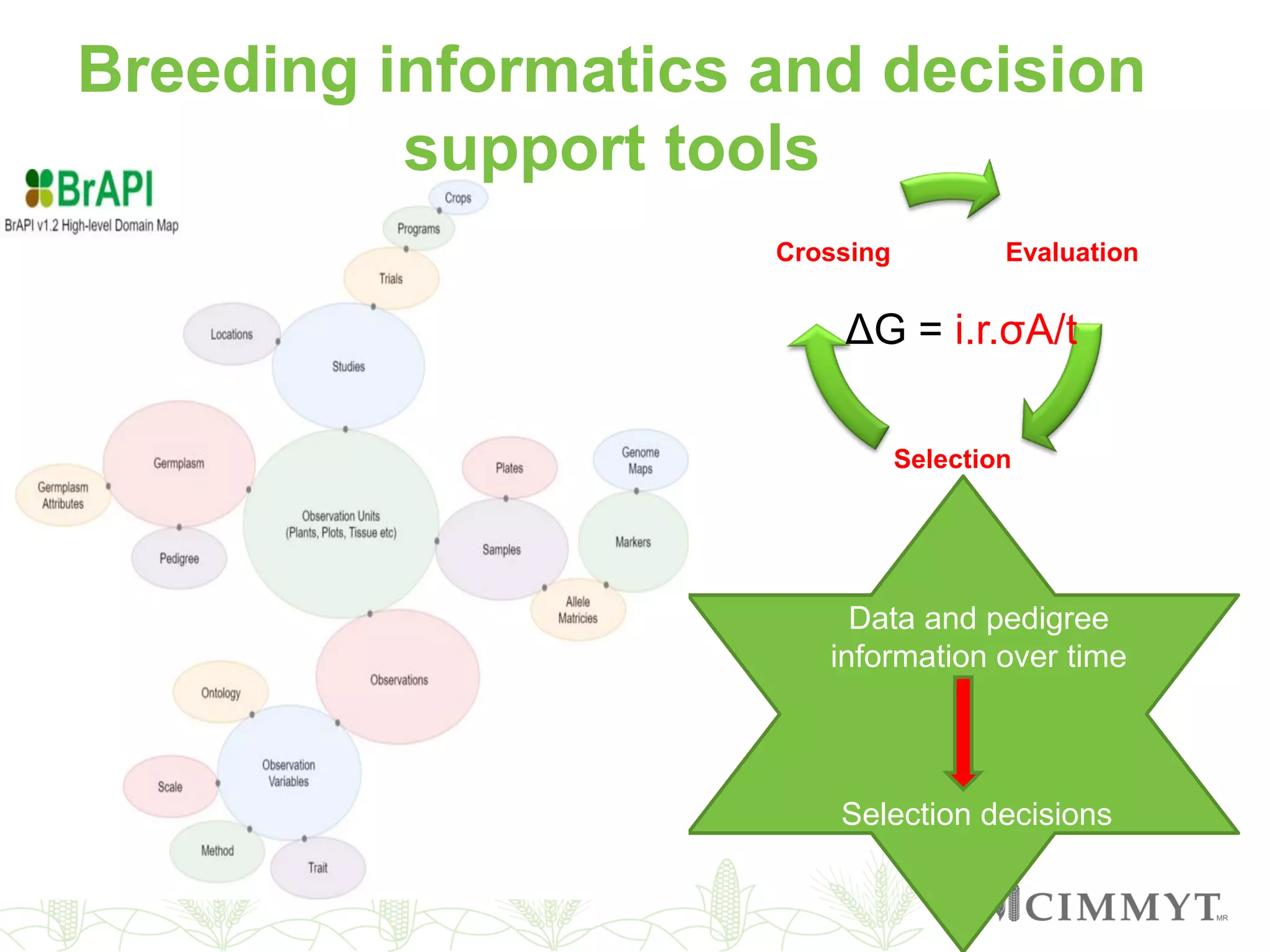
This document discusses the role of genomic and enabling technologies in enhancing genetic gains in maize breeding, particularly in tropical regions. It highlights the challenges and opportunities presented by maize genomics, including trait marker discovery, genomic prediction, and high-throughput genotyping. The document emphasizes the importance of integrating various genomic tools and breeding informatics to optimize breeding cycles and achieve better yield outcomes.