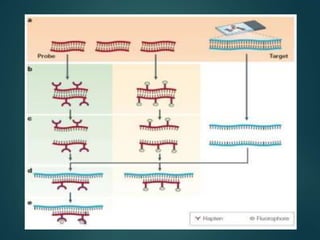
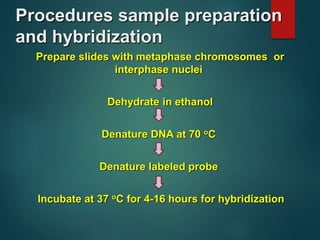
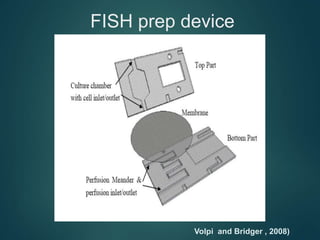
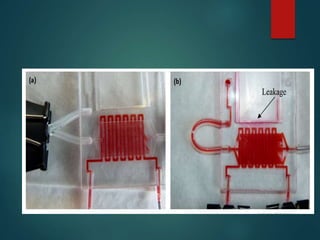
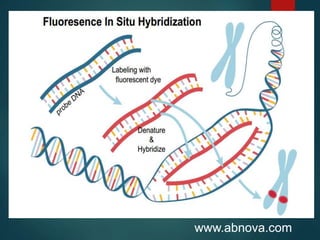
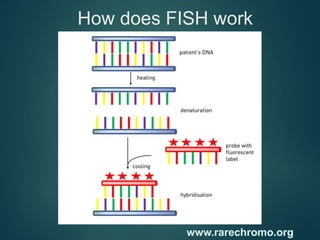
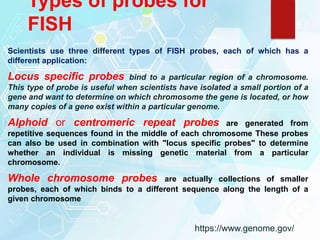
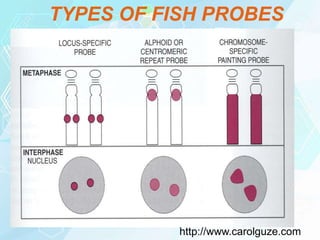
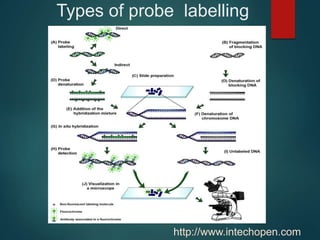
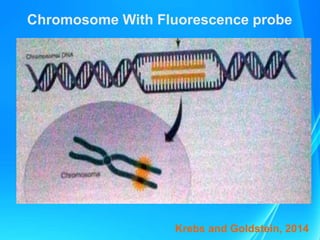
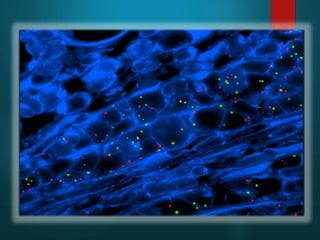
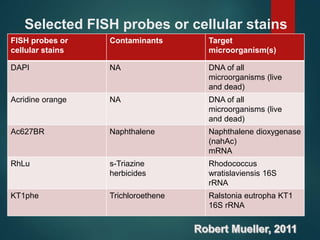
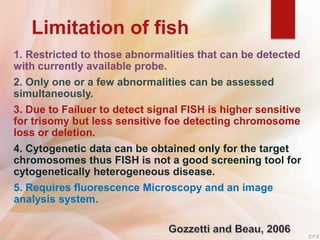
FISH is a technique that uses fluorescent probes that bind to complementary DNA sequences on chromosomes to detect chromosomal abnormalities. It allows detection of structural changes like deletions and duplications not visible under a microscope. FISH gained recognition supporting genome mapping efforts. It involves labeling probes, denaturing probe and target DNA, and allowing hybridization. Results are then viewed under a fluorescence microscope. FISH has applications in detecting abnormalities, identifying marker chromosomes, and monitoring therapy. It provides advantages over traditional cytogenetics like rapid analysis of non-dividing cells.