Embed presentation
Downloaded 65 times


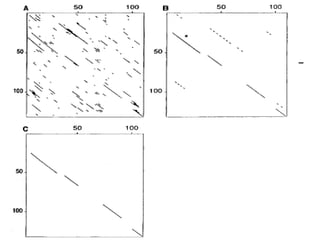

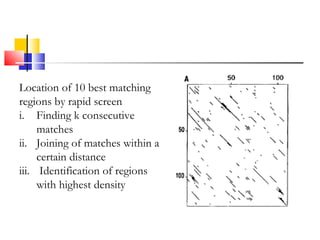
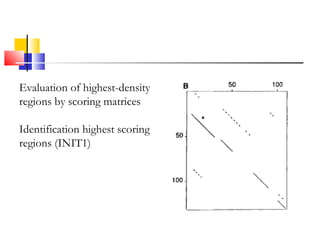
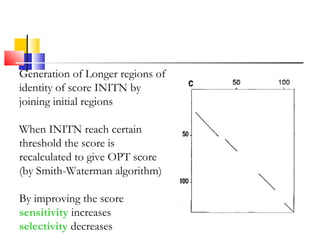

FASTA is a program for rapidly aligning protein and DNA sequences. It searches for matching k-tuples or sequence words and builds a local alignment based on these matches. It identifies the 10 best matching regions through k-tuple screening, joins nearby matches, and finds the highest density regions. It generates longer regions of identity and recalculates scores, with INITN and OPT scores used to rank database matches. Sensitivity refers to the ability to locate distantly related family members with limited similarity.









